Qiangwei Peng
WFR-MFM: One-Step Inference for Dynamic Unbalanced Optimal Transport
Jan 28, 2026Abstract:Reconstructing dynamical evolution from limited observations is a fundamental challenge in single-cell biology, where dynamic unbalanced optimal transport provides a principled framework for modeling coupled transport and mass variation. However, existing approaches rely on trajectory simulation at inference time, making inference a key bottleneck for scalable applications. In this work, we propose a mean-flow framework for unbalanced flow matching that summarizes both transport and mass-growth dynamics over arbitrary time intervals using mean velocity and mass-growth fields, enabling fast one-step generation without trajectory simulation. To solve dynamic unbalanced optimal transport under the Wasserstein-Fisher-Rao geometry, we further build on this framework to develop Wasserstein-Fisher-Rao Mean Flow Matching (WFR-MFM). Across synthetic and real single-cell RNA sequencing datasets, WFR-MFM achieves orders-of-magnitude faster inference than a range of existing baselines while maintaining high predictive accuracy, and enables efficient perturbation response prediction on large synthetic datasets with thousands of conditions.
WFR-FM: Simulation-Free Dynamic Unbalanced Optimal Transport
Jan 11, 2026Abstract:The Wasserstein-Fisher-Rao (WFR) metric extends dynamic optimal transport (OT) by coupling displacement with change of mass, providing a principled geometry for modeling unbalanced snapshot dynamics. Existing WFR solvers, however, are often unstable, computationally expensive, and difficult to scale. Here we introduce WFR Flow Matching (WFR-FM), a simulation-free training algorithm that unifies flow matching with dynamic unbalanced OT. Unlike classical flow matching which regresses only a transport vector field, WFR-FM simultaneously regresses a vector field for displacement and a scalar growth rate function for birth-death dynamics, yielding continuous flows under the WFR geometry. Theoretically, we show that minimizing the WFR-FM loss exactly recovers WFR geodesics. Empirically, WFR-FM yields more accurate and robust trajectory inference in single-cell biology, reconstructing consistent dynamics with proliferation and apoptosis, estimating time-varying growth fields, and applying to generative dynamics under imbalanced data. It outperforms state-of-the-art baselines in efficiency, stability, and reconstruction accuracy. Overall, WFR-FM establishes a unified and efficient paradigm for learning dynamical systems from unbalanced snapshots, where not only states but also mass evolve over time.
Integrating Dynamical Systems Modeling with Spatiotemporal scRNA-seq Data Analysis
Mar 14, 2025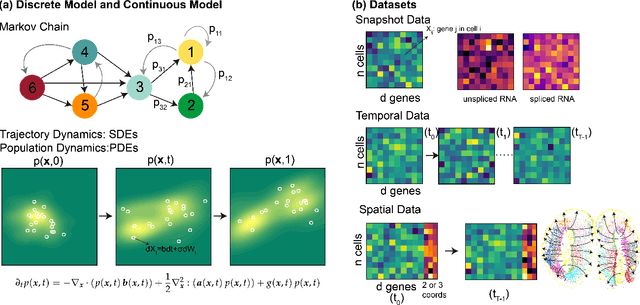
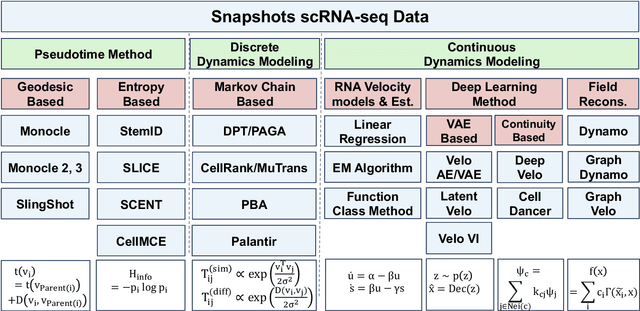
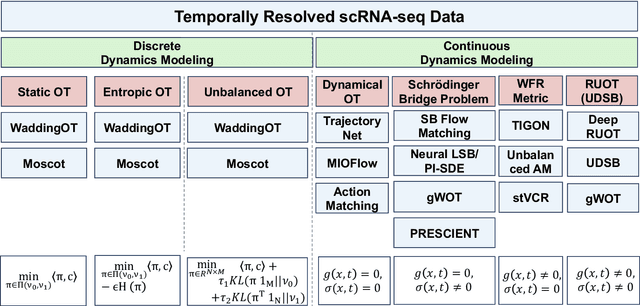
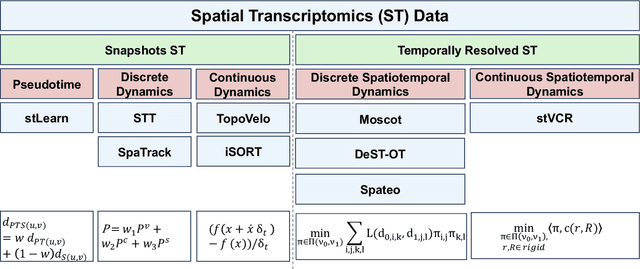
Abstract:Understanding the dynamic nature of biological systems is fundamental to deciphering cellular behavior, developmental processes, and disease progression. Single-cell RNA sequencing (scRNA-seq) has provided static snapshots of gene expression, offering valuable insights into cellular states at a single time point. Recent advancements in temporally resolved scRNA-seq, spatial transcriptomics (ST), and time-series spatial transcriptomics (temporal-ST) have further revolutionized our ability to study the spatiotemporal dynamics of individual cells. These technologies, when combined with computational frameworks such as Markov chains, stochastic differential equations (SDEs), and generative models like optimal transport and Schr\"odinger bridges, enable the reconstruction of dynamic cellular trajectories and cell fate decisions. This review discusses how these dynamical system approaches offer new opportunities to model and infer cellular dynamics from a systematic perspective.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge