Qiang Yue
FPL+: Filtered Pseudo Label-based Unsupervised Cross-Modality Adaptation for 3D Medical Image Segmentation
Apr 07, 2024



Abstract:Adapting a medical image segmentation model to a new domain is important for improving its cross-domain transferability, and due to the expensive annotation process, Unsupervised Domain Adaptation (UDA) is appealing where only unlabeled images are needed for the adaptation. Existing UDA methods are mainly based on image or feature alignment with adversarial training for regularization, and they are limited by insufficient supervision in the target domain. In this paper, we propose an enhanced Filtered Pseudo Label (FPL+)-based UDA method for 3D medical image segmentation. It first uses cross-domain data augmentation to translate labeled images in the source domain to a dual-domain training set consisting of a pseudo source-domain set and a pseudo target-domain set. To leverage the dual-domain augmented images to train a pseudo label generator, domain-specific batch normalization layers are used to deal with the domain shift while learning the domain-invariant structure features, generating high-quality pseudo labels for target-domain images. We then combine labeled source-domain images and target-domain images with pseudo labels to train a final segmentor, where image-level weighting based on uncertainty estimation and pixel-level weighting based on dual-domain consensus are proposed to mitigate the adverse effect of noisy pseudo labels. Experiments on three public multi-modal datasets for Vestibular Schwannoma, brain tumor and whole heart segmentation show that our method surpassed ten state-of-the-art UDA methods, and it even achieved better results than fully supervised learning in the target domain in some cases.
Routing-Guided Learned Product Quantization for Graph-Based Approximate Nearest Neighbor Search
Nov 30, 2023Abstract:Given a vector dataset $\mathcal{X}$, a query vector $\vec{x}_q$, graph-based Approximate Nearest Neighbor Search (ANNS) aims to build a proximity graph (PG) as an index of $\mathcal{X}$ and approximately return vectors with minimum distances to $\vec{x}_q$ by searching over the PG index. It suffers from the large-scale $\mathcal{X}$ because a PG with full vectors is too large to fit into the memory, e.g., a billion-scale $\mathcal{X}$ in 128 dimensions would consume nearly 600 GB memory. To solve this, Product Quantization (PQ) integrated graph-based ANNS is proposed to reduce the memory usage, using smaller compact codes of quantized vectors in memory instead of the large original vectors. Existing PQ methods do not consider the important routing features of PG, resulting in low-quality quantized vectors that affect the ANNS's effectiveness. In this paper, we present an end-to-end Routing-guided learned Product Quantization (RPQ) for graph-based ANNS. It consists of (1) a \textit{differentiable quantizer} used to make the standard discrete PQ differentiable to suit for back-propagation of end-to-end learning, (2) a \textit{sampling-based feature extractor} used to extract neighborhood and routing features of a PG, and (3) a \textit{multi-feature joint training module} with two types of feature-aware losses to continuously optimize the differentiable quantizer. As a result, the inherent features of a PG would be embedded into the learned PQ, generating high-quality quantized vectors. Moreover, we integrate our RPQ with the state-of-the-art DiskANN and existing popular PGs to improve their performance. Comprehensive experiments on real-world large-scale datasets (from 1M to 1B) demonstrate RPQ's superiority, e.g., 1.7$\times$-4.2$\times$ improvement on QPS at the same recall@10 of 95\%.
PA-Seg: Learning from Point Annotations for 3D Medical Image Segmentation using Contextual Regularization and Cross Knowledge Distillation
Aug 11, 2022



Abstract:The success of Convolutional Neural Networks (CNNs) in 3D medical image segmentation relies on massive fully annotated 3D volumes for training that are time-consuming and labor-intensive to acquire. In this paper, we propose to annotate a segmentation target with only seven points in 3D medical images, and design a two-stage weakly supervised learning framework PA-Seg. In the first stage, we employ geodesic distance transform to expand the seed points to provide more supervision signal. To further deal with unannotated image regions during training, we propose two contextual regularization strategies, i.e., multi-view Conditional Random Field (mCRF) loss and Variance Minimization (VM) loss, where the first one encourages pixels with similar features to have consistent labels, and the second one minimizes the intensity variance for the segmented foreground and background, respectively. In the second stage, we use predictions obtained by the model pre-trained in the first stage as pseudo labels. To overcome noises in the pseudo labels, we introduce a Self and Cross Monitoring (SCM) strategy, which combines self-training with Cross Knowledge Distillation (CKD) between a primary model and an auxiliary model that learn from soft labels generated by each other. Experiments on public datasets for Vestibular Schwannoma (VS) segmentation and Brain Tumor Segmentation (BraTS) demonstrated that our model trained in the first stage outperforms existing state-of-the-art weakly supervised approaches by a large margin, and after using SCM for additional training, the model can achieve competitive performance compared with the fully supervised counterpart on the BraTS dataset.
Navigable Proximity Graph-Driven Native Hybrid Queries with Structured and Unstructured Constraints
Mar 25, 2022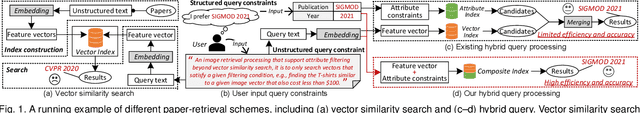
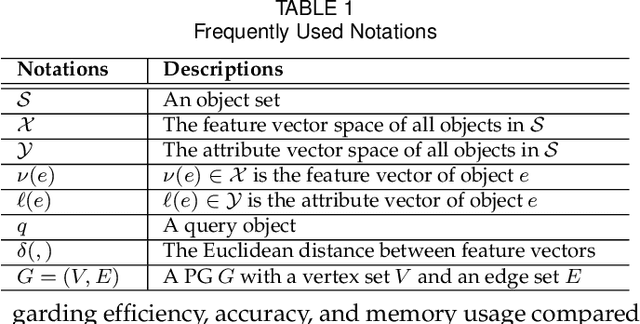
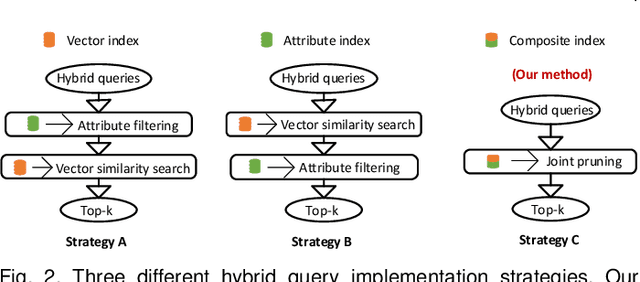
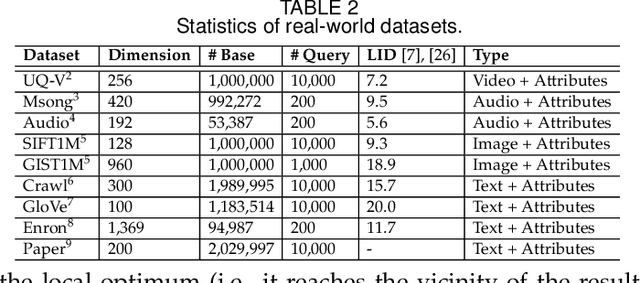
Abstract:As research interest surges, vector similarity search is applied in multiple fields, including data mining, computer vision, and information retrieval. {Given a set of objects (e.g., a set of images) and a query object, we can easily transform each object into a feature vector and apply the vector similarity search to retrieve the most similar objects. However, the original vector similarity search cannot well support \textit{hybrid queries}, where users not only input unstructured query constraint (i.e., the feature vector of query object) but also structured query constraint (i.e., the desired attributes of interest). Hybrid query processing aims at identifying these objects with similar feature vectors to query object and satisfying the given attribute constraints. Recent efforts have attempted to answer a hybrid query by performing attribute filtering and vector similarity search separately and then merging the results later, which limits efficiency and accuracy because they are not purpose-built for hybrid queries.} In this paper, we propose a native hybrid query (NHQ) framework based on proximity graph (PG), which provides the specialized \textit{composite index and joint pruning} modules for hybrid queries. We easily deploy existing various PGs on this framework to process hybrid queries efficiently. Moreover, we present two novel navigable PGs (NPGs) with optimized edge selection and routing strategies, which obtain better overall performance than existing PGs. After that, we deploy the proposed NPGs in NHQ to form two hybrid query methods, which significantly outperform the state-of-the-art competitors on all experimental datasets (10$\times$ faster under the same \textit{Recall}), including eight public and one in-house real-world datasets. Our code and datasets have been released at \url{https://github.com/AshenOn3/NHQ}.
A Comprehensive Survey and Experimental Comparison of Graph-Based Approximate Nearest Neighbor Search
Jan 29, 2021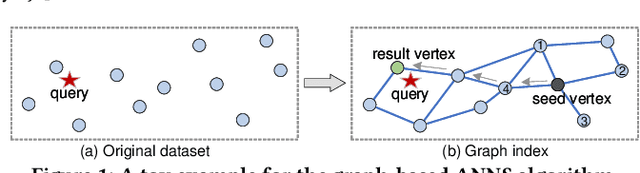
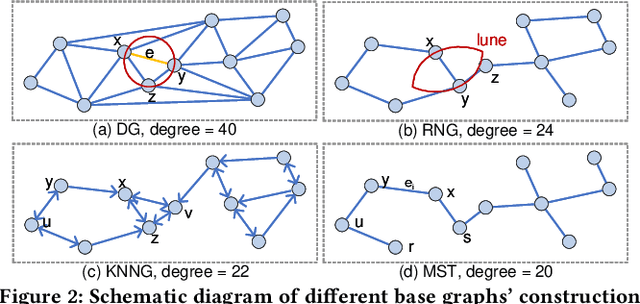
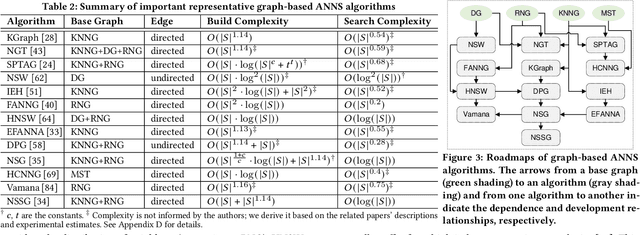
Abstract:Approximate nearest neighbor search (ANNS) constitutes an important operation in a multitude of applications, including recommendation systems, information retrieval, and pattern recognition. In the past decade, graph-based ANNS algorithms have been the leading paradigm in this domain, with dozens of graph-based ANNS algorithms proposed. Such algorithms aim to provide effective, efficient solutions for retrieving the nearest neighbors for a given query. Nevertheless, these efforts focus on developing and optimizing algorithms with different approaches, so there is a real need for a comprehensive survey about the approaches' relative performance, strengths, and pitfalls. Thus here we provide a thorough comparative analysis and experimental evaluation of 13 representative graph-based ANNS algorithms via a new taxonomy and fine-grained pipeline. We compared each algorithm in a uniform test environment on eight real-world datasets and 12 synthetic datasets with varying sizes and characteristics. Our study yields novel discoveries, offerings several useful principles to improve algorithms. This effort also helped us pinpoint algorithms' working portions, along with rule-of-thumb recommendations about promising research directions and suitable algorithms for practitioners in different fields.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge