Pratik Kayal
Tables to LaTeX: structure and content extraction from scientific tables
Oct 31, 2022Abstract:Scientific documents contain tables that list important information in a concise fashion. Structure and content extraction from tables embedded within PDF research documents is a very challenging task due to the existence of visual features like spanning cells and content features like mathematical symbols and equations. Most existing table structure identification methods tend to ignore these academic writing features. In this paper, we adapt the transformer-based language modeling paradigm for scientific table structure and content extraction. Specifically, the proposed model converts a tabular image to its corresponding LaTeX source code. Overall, we outperform the current state-of-the-art baselines and achieve an exact match accuracy of 70.35 and 49.69% on table structure and content extraction, respectively. Further analysis demonstrates that the proposed models efficiently identify the number of rows and columns, the alphanumeric characters, the LaTeX tokens, and symbols.
On Adversarial Robustness of Synthetic Code Generation
Jun 22, 2021
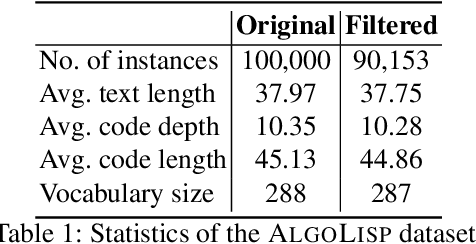
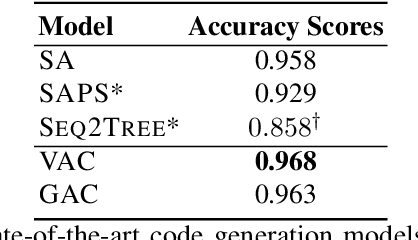

Abstract:Automatic code synthesis from natural language descriptions is a challenging task. We witness massive progress in developing code generation systems for domain-specific languages (DSLs) employing sequence-to-sequence deep learning techniques in the recent past. In this paper, we specifically experiment with \textsc{AlgoLisp} DSL-based generative models and showcase the existence of significant dataset bias through different classes of adversarial examples. We also experiment with two variants of Transformer-based models that outperform all existing \textsc{AlgoLisp} DSL-based code generation baselines. Consistent with the current state-of-the-art systems, our proposed models, too, achieve poor performance under adversarial settings. Therefore, we propose several dataset augmentation techniques to reduce bias and showcase their efficacy using robust experimentation.
ICDAR 2021 Competition on Scientific Table Image Recognition to LaTeX
May 30, 2021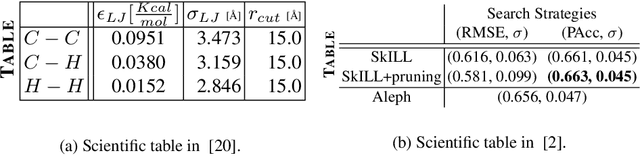
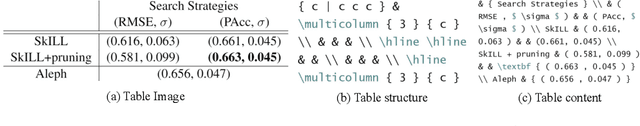

Abstract:Tables present important information concisely in many scientific documents. Visual features like mathematical symbols, equations, and spanning cells make structure and content extraction from tables embedded in research documents difficult. This paper discusses the dataset, tasks, participants' methods, and results of the ICDAR 2021 Competition on Scientific Table Image Recognition to LaTeX. Specifically, the task of the competition is to convert a tabular image to its corresponding LaTeX source code. We proposed two subtasks. In Subtask 1, we ask the participants to reconstruct the LaTeX structure code from an image. In Subtask 2, we ask the participants to reconstruct the LaTeX content code from an image. This report describes the datasets and ground truth specification, details the performance evaluation metrics used, presents the final results, and summarizes the participating methods. Submission by team VCGroup got the highest Exact Match accuracy score of 74% for Subtask 1 and 55% for Subtask 2, beating previous baselines by 5% and 12%, respectively. Although improvements can still be made to the recognition capabilities of models, this competition contributes to the development of fully automated table recognition systems by challenging practitioners to solve problems under specific constraints and sharing their approaches; the platform will remain available for post-challenge submissions at https://competitions.codalab.org/competitions/26979 .
* Competition on Scientific Table Image Recognition to LaTeX
TabLeX: A Benchmark Dataset for Structure and Content Information Extraction from Scientific Tables
May 12, 2021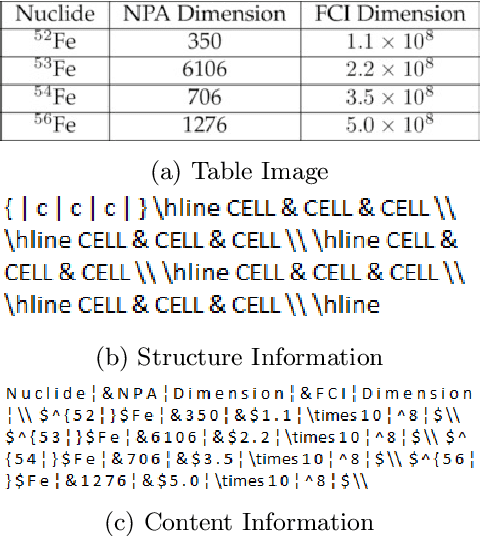
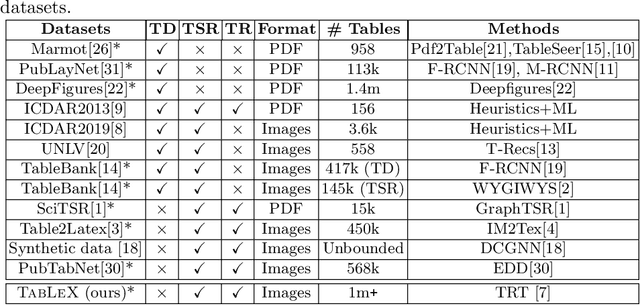
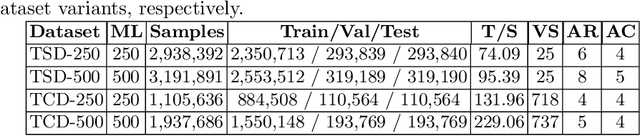
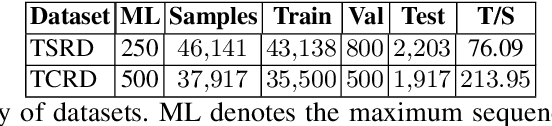
Abstract:Information Extraction (IE) from the tables present in scientific articles is challenging due to complicated tabular representations and complex embedded text. This paper presents TabLeX, a large-scale benchmark dataset comprising table images generated from scientific articles. TabLeX consists of two subsets, one for table structure extraction and the other for table content extraction. Each table image is accompanied by its corresponding LATEX source code. To facilitate the development of robust table IE tools, TabLeX contains images in different aspect ratios and in a variety of fonts. Our analysis sheds light on the shortcomings of current state-of-the-art table extraction models and shows that they fail on even simple table images. Towards the end, we experiment with a transformer-based existing baseline to report performance scores. In contrast to the static benchmarks, we plan to augment this dataset with more complex and diverse tables at regular intervals.
Weakly-Supervised Deep Learning for Domain Invariant Sentiment Classification
Nov 23, 2019



Abstract:The task of learning a sentiment classification model that adapts well to any target domain, different from the source domain, is a challenging problem. Majority of the existing approaches focus on learning a common representation by leveraging both source and target data during training. In this paper, we introduce a two-stage training procedure that leverages weakly supervised datasets for developing simple lift-and-shift-based predictive models without being exposed to the target domain during the training phase. Experimental results show that transfer with weak supervision from a source domain to various target domains provides performance very close to that obtained via supervised training on the target domain itself.
PlantDoc: A Dataset for Visual Plant Disease Detection
Nov 23, 2019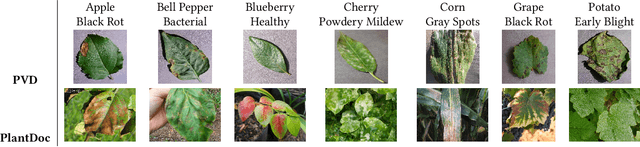

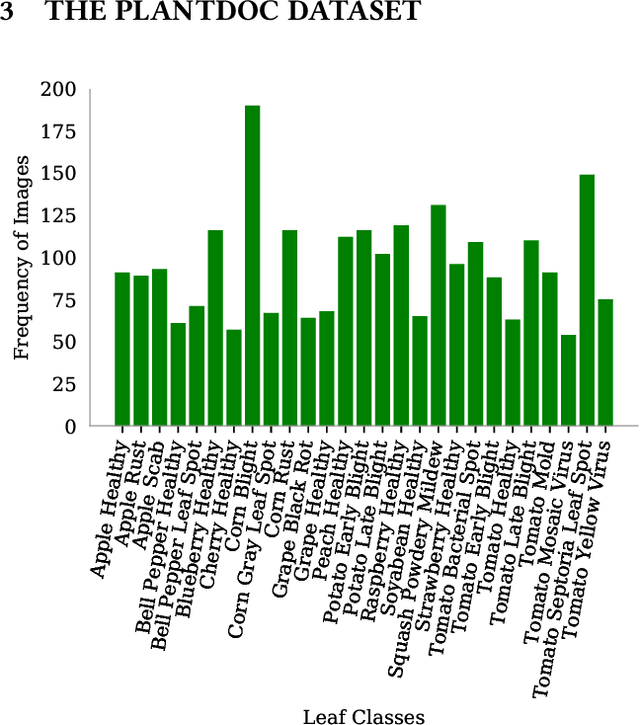

Abstract:India loses 35% of the annual crop yield due to plant diseases. Early detection of plant diseases remains difficult due to the lack of lab infrastructure and expertise. In this paper, we explore the possibility of computer vision approaches for scalable and early plant disease detection. The lack of availability of sufficiently large-scale non-lab data set remains a major challenge for enabling vision based plant disease detection. Against this background, we present PlantDoc: a dataset for visual plant disease detection. Our dataset contains 2,598 data points in total across 13 plant species and up to 17 classes of diseases, involving approximately 300 human hours of effort in annotating internet scraped images. To show the efficacy of our dataset, we learn 3 models for the task of plant disease classification. Our results show that modelling using our dataset can increase the classification accuracy by up to 31%. We believe that our dataset can help reduce the entry barrier of computer vision techniques in plant disease detection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge