Philipp Singer
H2O-Danube3 Technical Report
Jul 12, 2024



Abstract:We present H2O-Danube3, a series of small language models consisting of H2O-Danube3-4B, trained on 6T tokens and H2O-Danube3-500M, trained on 4T tokens. Our models are pre-trained on high quality Web data consisting of primarily English tokens in three stages with different data mixes before final supervised tuning for chat version. The models exhibit highly competitive metrics across a multitude of academic, chat, and fine-tuning benchmarks. Thanks to its compact architecture, H2O-Danube3 can be efficiently run on a modern smartphone, enabling local inference and rapid processing capabilities even on mobile devices. We make all models openly available under Apache 2.0 license further democratizing LLMs to a wider audience economically.
H2O-Danube-1.8B Technical Report
Jan 30, 2024Abstract:We present H2O-Danube-1.8B, a 1.8B language model trained on 1T tokens following the core principles of LLama 2 and Mistral. We leverage and refine various techniques for pre-training large language models. Although our model is trained on significantly fewer total tokens compared to reference models of similar size, it exhibits highly competitive metrics across a multitude of benchmarks. We additionally release a chat model trained with supervised fine-tuning followed by direct preference optimization. We make H2O-Danube-1.8B openly available under Apache 2.0 license further democratizing LLMs to a wider audience economically.
H2O Open Ecosystem for State-of-the-art Large Language Models
Oct 23, 2023



Abstract:Large Language Models (LLMs) represent a revolution in AI. However, they also pose many significant risks, such as the presence of biased, private, copyrighted or harmful text. For this reason we need open, transparent and safe solutions. We introduce a complete open-source ecosystem for developing and testing LLMs. The goal of this project is to boost open alternatives to closed-source approaches. We release h2oGPT, a family of fine-tuned LLMs of diverse sizes. We also introduce H2O LLM Studio, a framework and no-code GUI designed for efficient fine-tuning, evaluation, and deployment of LLMs using the most recent state-of-the-art techniques. Our code and models are fully open-source. We believe this work helps to boost AI development and make it more accessible, efficient and trustworthy. The demo is available at: https://gpt.h2o.ai/
h2oGPT: Democratizing Large Language Models
Jun 16, 2023


Abstract:Applications built on top of Large Language Models (LLMs) such as GPT-4 represent a revolution in AI due to their human-level capabilities in natural language processing. However, they also pose many significant risks such as the presence of biased, private, or harmful text, and the unauthorized inclusion of copyrighted material. We introduce h2oGPT, a suite of open-source code repositories for the creation and use of LLMs based on Generative Pretrained Transformers (GPTs). The goal of this project is to create the world's best truly open-source alternative to closed-source approaches. In collaboration with and as part of the incredible and unstoppable open-source community, we open-source several fine-tuned h2oGPT models from 7 to 40 Billion parameters, ready for commercial use under fully permissive Apache 2.0 licenses. Included in our release is 100\% private document search using natural language. Open-source language models help boost AI development and make it more accessible and trustworthy. They lower entry hurdles, allowing people and groups to tailor these models to their needs. This openness increases innovation, transparency, and fairness. An open-source strategy is needed to share AI benefits fairly, and H2O.ai will continue to democratize AI and LLMs.
Recognizing bird species in diverse soundscapes under weak supervision
Jul 16, 2021



Abstract:We present a robust classification approach for avian vocalization in complex and diverse soundscapes, achieving second place in the BirdCLEF2021 challenge. We illustrate how to make full use of pre-trained convolutional neural networks, by using an efficient modeling and training routine supplemented by novel augmentation methods. Thereby, we improve the generalization of weakly labeled crowd-sourced data to productive data collected by autonomous recording units. As such, we illustrate how to progress towards an accurate automated assessment of avian population which would enable global biodiversity monitoring at scale, impossible by manual annotation.
Supporting large-scale image recognition with out-of-domain samples
Oct 04, 2020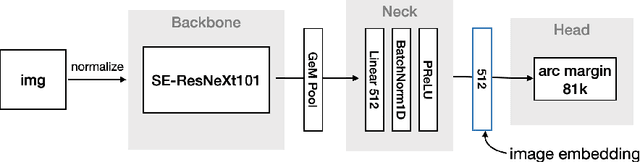
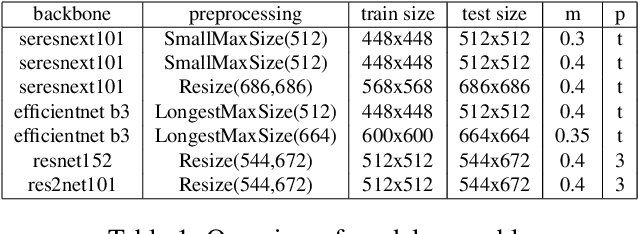

Abstract:This article presents an efficient end-to-end method to perform instance-level recognition employed to the task of labeling and ranking landmark images. In a first step, we embed images in a high dimensional feature space using convolutional neural networks trained with an additive angular margin loss and classify images using visual similarity. We then efficiently re-rank predictions and filter noise utilizing similarity to out-of-domain images. Using this approach we achieved the 1st place in the 2020 edition of the Google Landmark Recognition challenge.
Backtesting the predictability of COVID-19
Jul 22, 2020



Abstract:The advent of the COVID-19 pandemic has instigated unprecedented changes in many countries around the globe, putting a significant burden on the health sectors, affecting the macro economic conditions, and altering social interactions amongst the population. In response, the academic community has produced multiple forecasting models, approaches and algorithms to best predict the different indicators of COVID-19, such as the number of confirmed infected cases. Yet, researchers had little to no historical information about the pandemic at their disposal in order to inform their forecasting methods. Our work studies the predictive performance of models at various stages of the pandemic to better understand their fundamental uncertainty and the impact of data availability on such forecasts. We use historical data of COVID-19 infections from 253 regions from the period of 22nd January 2020 until 22nd June 2020 to predict, through a rolling window backtesting framework, the cumulative number of infected cases for the next 7 and 28 days. We implement three simple models to track the root mean squared logarithmic error in this 6-month span, a baseline model that always predicts the last known value of the cumulative confirmed cases, a power growth model and an epidemiological model called SEIRD. Prediction errors are substantially higher in early stages of the pandemic, resulting from limited data. Throughout the course of the pandemic, errors regress slowly, but steadily. The more confirmed cases a country exhibits at any point in time, the lower the error in forecasting future confirmed cases. We emphasize the significance of having a rigorous backtesting framework to accurately assess the predictive power of such models at any point in time during the outbreak which in turn can be used to assign the right level of certainty to these forecasts and facilitate better planning.
Discovering Beaten Paths in Collaborative Ontology-Engineering Projects using Markov Chains
Feb 29, 2016



Abstract:Biomedical taxonomies, thesauri and ontologies in the form of the International Classification of Diseases (ICD) as a taxonomy or the National Cancer Institute Thesaurus as an OWL-based ontology, play a critical role in acquiring, representing and processing information about human health. With increasing adoption and relevance, biomedical ontologies have also significantly increased in size. For example, the 11th revision of the ICD, which is currently under active development by the WHO contains nearly 50,000 classes representing a vast variety of different diseases and causes of death. This evolution in terms of size was accompanied by an evolution in the way ontologies are engineered. Because no single individual has the expertise to develop such large-scale ontologies, ontology-engineering projects have evolved from small-scale efforts involving just a few domain experts to large-scale projects that require effective collaboration between dozens or even hundreds of experts, practitioners and other stakeholders. Understanding how these stakeholders collaborate will enable us to improve editing environments that support such collaborations. We uncover how large ontology-engineering projects, such as the ICD in its 11th revision, unfold by analyzing usage logs of five different biomedical ontology-engineering projects of varying sizes and scopes using Markov chains. We discover intriguing interaction patterns (e.g., which properties users subsequently change) that suggest that large collaborative ontology-engineering projects are governed by a few general principles that determine and drive development. From our analysis, we identify commonalities and differences between different projects that have implications for project managers, ontology editors, developers and contributors working on collaborative ontology-engineering projects and tools in the biomedical domain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge