Peter Kraft
FastImpute: A Baseline for Open-source, Reference-Free Genotype Imputation Methods -- A Case Study in PRS313
Jul 12, 2024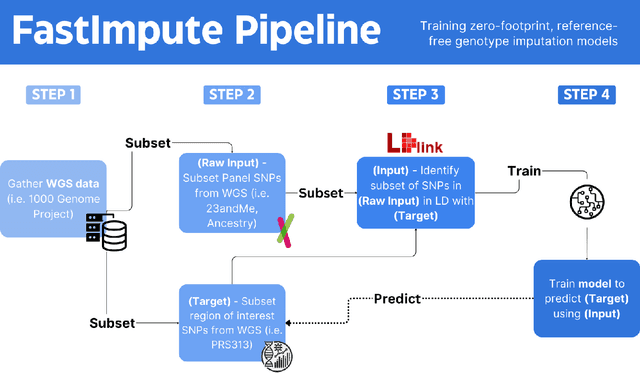
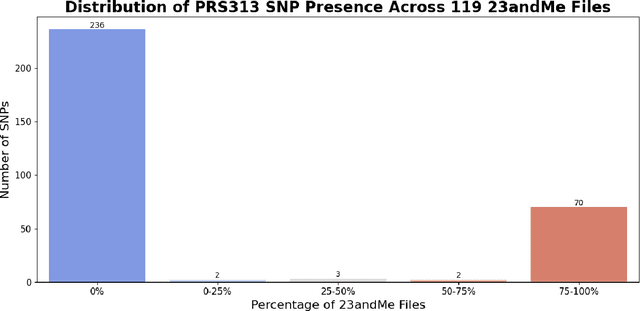
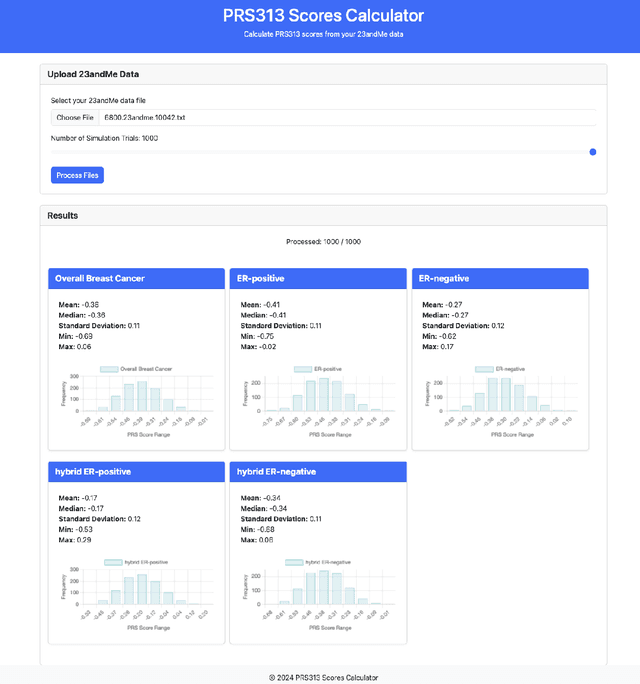
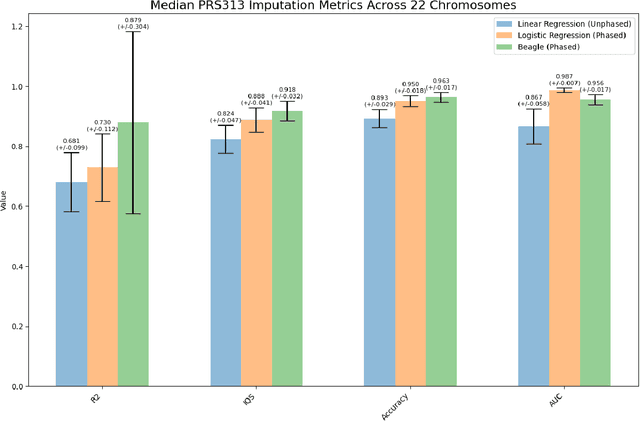
Abstract:Genotype imputation enhances genetic data by predicting missing SNPs using reference haplotype information. Traditional methods leverage linkage disequilibrium (LD) to infer untyped SNP genotypes, relying on the similarity of LD structures between genotyped target sets and fully sequenced reference panels. Recently, reference-free deep learning-based methods have emerged, offering a promising alternative by predicting missing genotypes without external databases, thereby enhancing privacy and accessibility. However, these methods often produce models with tens of millions of parameters, leading to challenges such as the need for substantial computational resources to train and inefficiency for client-sided deployment. Our study addresses these limitations by introducing a baseline for a novel genotype imputation pipeline that supports client-sided imputation models generalizable across any genotyping chip and genomic region. This approach enhances patient privacy by performing imputation directly on edge devices. As a case study, we focus on PRS313, a polygenic risk score comprising 313 SNPs used for breast cancer risk prediction. Utilizing consumer genetic panels such as 23andMe, our model democratizes access to personalized genetic insights by allowing 23andMe users to obtain their PRS313 score. We demonstrate that simple linear regression can significantly improve the accuracy of PRS313 scores when calculated using SNPs imputed from consumer gene panels, such as 23andMe. Our linear regression model achieved an R^2 of 0.86, compared to 0.33 without imputation and 0.28 with simple imputation (substituting missing SNPs with the minor allele frequency). These findings suggest that popular SNP analysis libraries could benefit from integrating linear regression models for genotype imputation, providing a viable and light-weight alternative to reference based imputation.
ACORN: Performant and Predicate-Agnostic Search Over Vector Embeddings and Structured Data
Mar 07, 2024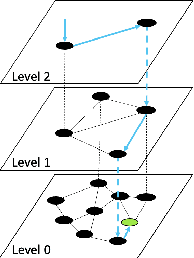
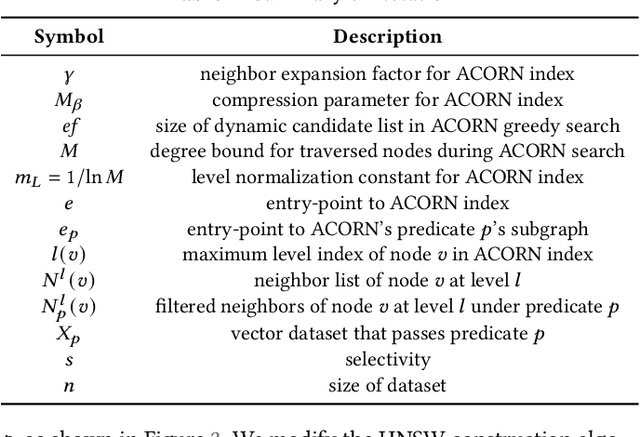
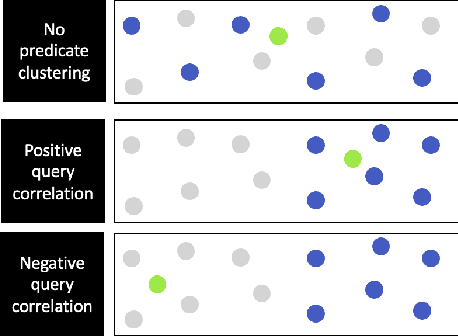
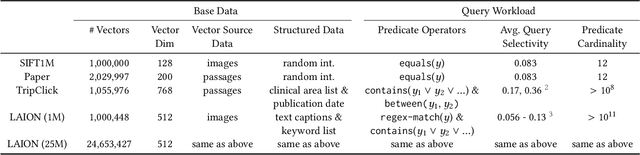
Abstract:Applications increasingly leverage mixed-modality data, and must jointly search over vector data, such as embedded images, text and video, as well as structured data, such as attributes and keywords. Proposed methods for this hybrid search setting either suffer from poor performance or support a severely restricted set of search predicates (e.g., only small sets of equality predicates), making them impractical for many applications. To address this, we present ACORN, an approach for performant and predicate-agnostic hybrid search. ACORN builds on Hierarchical Navigable Small Worlds (HNSW), a state-of-the-art graph-based approximate nearest neighbor index, and can be implemented efficiently by extending existing HNSW libraries. ACORN introduces the idea of predicate subgraph traversal to emulate a theoretically ideal, but impractical, hybrid search strategy. ACORN's predicate-agnostic construction algorithm is designed to enable this effective search strategy, while supporting a wide array of predicate sets and query semantics. We systematically evaluate ACORN on both prior benchmark datasets, with simple, low-cardinality predicate sets, and complex multi-modal datasets not supported by prior methods. We show that ACORN achieves state-of-the-art performance on all datasets, outperforming prior methods with 2-1,000x higher throughput at a fixed recall.
Machine Learning with DBOS
Aug 10, 2022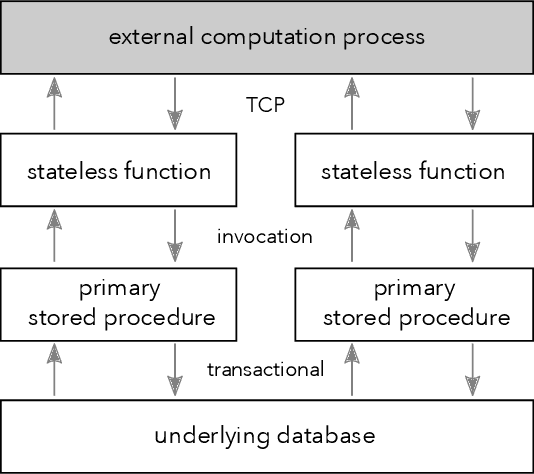
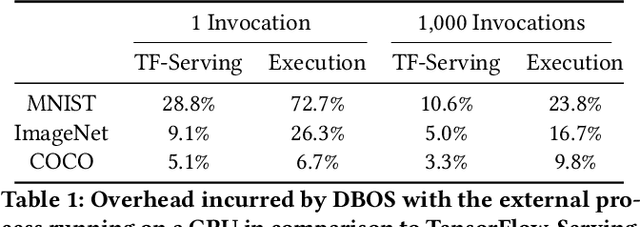

Abstract:We recently proposed a new cluster operating system stack, DBOS, centered on a DBMS. DBOS enables unique support for ML applications by encapsulating ML code within stored procedures, centralizing ancillary ML data, providing security built into the underlying DBMS, co-locating ML code and data, and tracking data and workflow provenance. Here we demonstrate a subset of these benefits around two ML applications. We first show that image classification and object detection models using GPUs can be served as DBOS stored procedures with performance competitive to existing systems. We then present a 1D CNN trained to detect anomalies in HTTP requests on DBOS-backed web services, achieving SOTA results. We use this model to develop an interactive anomaly detection system and evaluate it through qualitative user feedback, demonstrating its usefulness as a proof of concept for future work to develop learned real-time security services on top of DBOS.
Willump: A Statistically-Aware End-to-end Optimizer for Machine Learning Inference
Jun 03, 2019
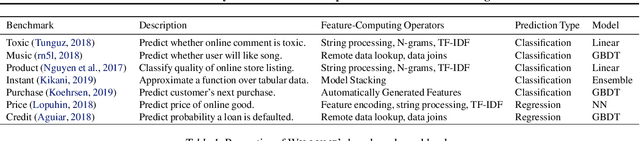
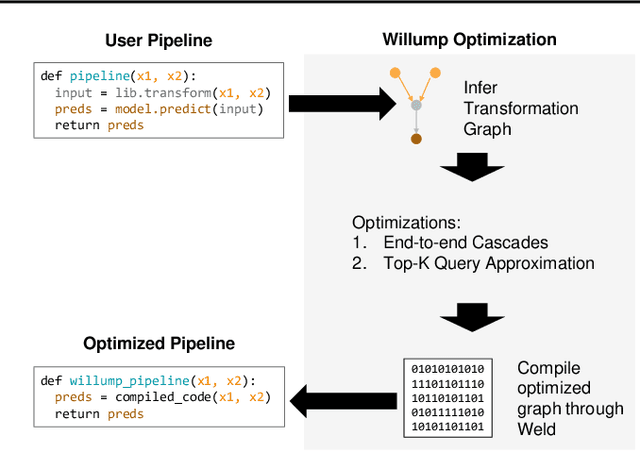
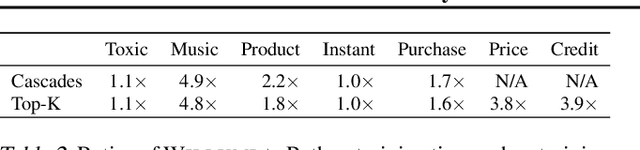
Abstract:Machine learning (ML) has become increasingly important and performance-critical in modern data centers. This has led to interest in model serving systems, which perform ML inference and serve predictions to end-user applications. However, most existing model serving systems approach ML inference as an extension of conventional data serving workloads and miss critical opportunities for performance. In this paper, we present Willump, a statistically-aware optimizer for ML inference that takes advantage of key properties of ML inference not shared by traditional workloads. First, ML models can often be approximated efficiently on many "easy" inputs by judiciously using a less expensive model for these inputs (e.g., not computing all the input features). Willump automatically generates such approximations from an ML inference pipeline, providing up to 4.1$\times$ speedup without statistically significant accuracy loss. Second, ML models are often used in higher-level end-to-end queries in an ML application, such as computing the top K predictions for a recommendation model. Willump optimizes inference based on these higher-level queries by up to 5.7$\times$ over na\"ive batch inference. Willump combines these novel optimizations with standard compiler optimizations and a computation graph-aware feature caching scheme to automatically generate fast inference code for ML pipelines. We show that Willump improves performance of real-world ML inference pipelines by up to 23$\times$, with its novel optimizations giving 3.6-5.7$\times$ speedups over compilation. We also show that Willump integrates easily with existing model serving systems, such as Clipper.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge