FastImpute: A Baseline for Open-source, Reference-Free Genotype Imputation Methods -- A Case Study in PRS313
Paper and Code
Jul 12, 2024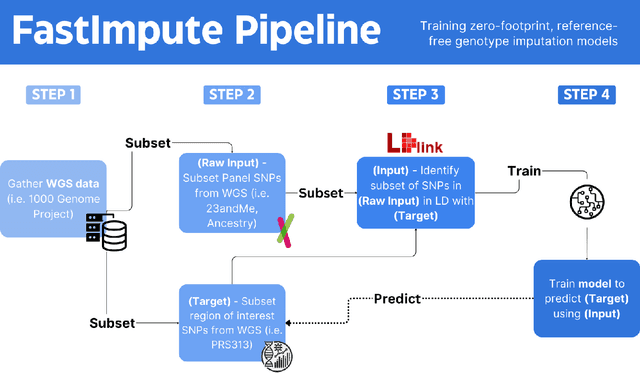
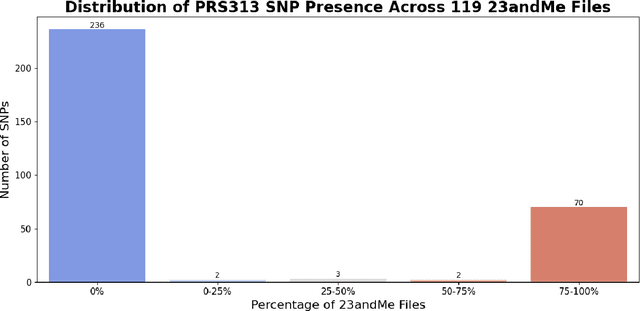
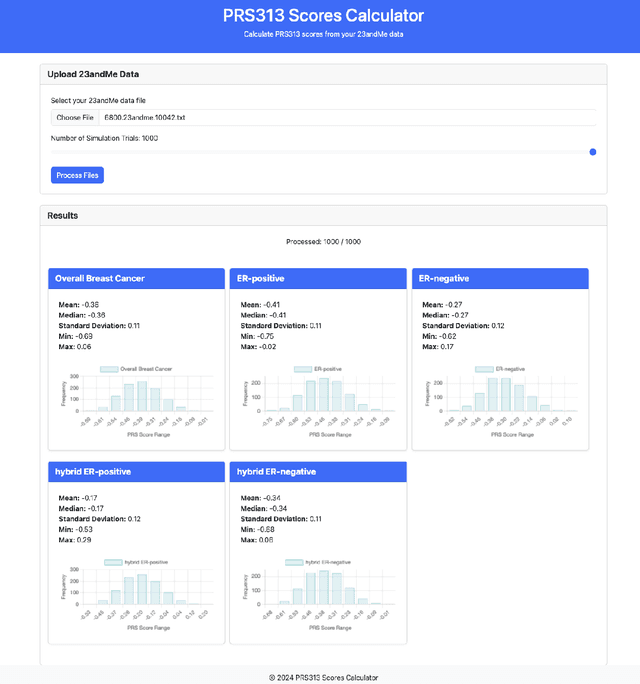
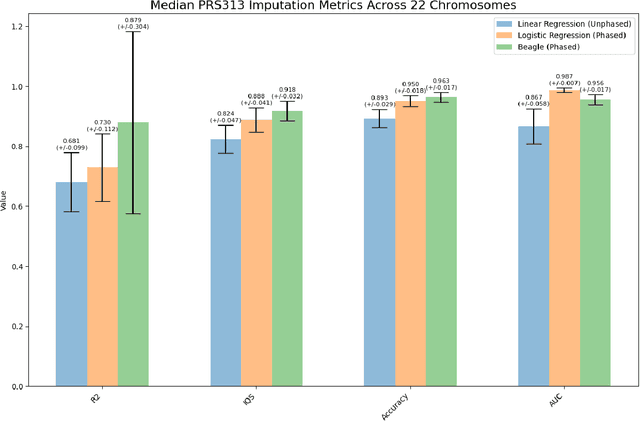
Genotype imputation enhances genetic data by predicting missing SNPs using reference haplotype information. Traditional methods leverage linkage disequilibrium (LD) to infer untyped SNP genotypes, relying on the similarity of LD structures between genotyped target sets and fully sequenced reference panels. Recently, reference-free deep learning-based methods have emerged, offering a promising alternative by predicting missing genotypes without external databases, thereby enhancing privacy and accessibility. However, these methods often produce models with tens of millions of parameters, leading to challenges such as the need for substantial computational resources to train and inefficiency for client-sided deployment. Our study addresses these limitations by introducing a baseline for a novel genotype imputation pipeline that supports client-sided imputation models generalizable across any genotyping chip and genomic region. This approach enhances patient privacy by performing imputation directly on edge devices. As a case study, we focus on PRS313, a polygenic risk score comprising 313 SNPs used for breast cancer risk prediction. Utilizing consumer genetic panels such as 23andMe, our model democratizes access to personalized genetic insights by allowing 23andMe users to obtain their PRS313 score. We demonstrate that simple linear regression can significantly improve the accuracy of PRS313 scores when calculated using SNPs imputed from consumer gene panels, such as 23andMe. Our linear regression model achieved an R^2 of 0.86, compared to 0.33 without imputation and 0.28 with simple imputation (substituting missing SNPs with the minor allele frequency). These findings suggest that popular SNP analysis libraries could benefit from integrating linear regression models for genotype imputation, providing a viable and light-weight alternative to reference based imputation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge