Pengfei Luo
Look As You Think: Unifying Reasoning and Visual Evidence Attribution for Verifiable Document RAG via Reinforcement Learning
Nov 15, 2025Abstract:Aiming to identify precise evidence sources from visual documents, visual evidence attribution for visual document retrieval-augmented generation (VD-RAG) ensures reliable and verifiable predictions from vision-language models (VLMs) in multimodal question answering. Most existing methods adopt end-to-end training to facilitate intuitive answer verification. However, they lack fine-grained supervision and progressive traceability throughout the reasoning process. In this paper, we introduce the Chain-of-Evidence (CoE) paradigm for VD-RAG. CoE unifies Chain-of-Thought (CoT) reasoning and visual evidence attribution by grounding reference elements in reasoning steps to specific regions with bounding boxes and page indexes. To enable VLMs to generate such evidence-grounded reasoning, we propose Look As You Think (LAT), a reinforcement learning framework that trains models to produce verifiable reasoning paths with consistent attribution. During training, LAT evaluates the attribution consistency of each evidence region and provides rewards only when the CoE trajectory yields correct answers, encouraging process-level self-verification. Experiments on vanilla Qwen2.5-VL-7B-Instruct with Paper- and Wiki-VISA benchmarks show that LAT consistently improves the vanilla model in both single- and multi-image settings, yielding average gains of 8.23% in soft exact match (EM) and 47.0% in IoU@0.5. Meanwhile, LAT not only outperforms the supervised fine-tuning baseline, which is trained to directly produce answers with attribution, but also exhibits stronger generalization across domains.
ImageScope: Unifying Language-Guided Image Retrieval via Large Multimodal Model Collective Reasoning
Mar 13, 2025



Abstract:With the proliferation of images in online content, language-guided image retrieval (LGIR) has emerged as a research hotspot over the past decade, encompassing a variety of subtasks with diverse input forms. While the development of large multimodal models (LMMs) has significantly facilitated these tasks, existing approaches often address them in isolation, requiring the construction of separate systems for each task. This not only increases system complexity and maintenance costs, but also exacerbates challenges stemming from language ambiguity and complex image content, making it difficult for retrieval systems to provide accurate and reliable results. To this end, we propose ImageScope, a training-free, three-stage framework that leverages collective reasoning to unify LGIR tasks. The key insight behind the unification lies in the compositional nature of language, which transforms diverse LGIR tasks into a generalized text-to-image retrieval process, along with the reasoning of LMMs serving as a universal verification to refine the results. To be specific, in the first stage, we improve the robustness of the framework by synthesizing search intents across varying levels of semantic granularity using chain-of-thought (CoT) reasoning. In the second and third stages, we then reflect on retrieval results by verifying predicate propositions locally, and performing pairwise evaluations globally. Experiments conducted on six LGIR datasets demonstrate that ImageScope outperforms competitive baselines. Comprehensive evaluations and ablation studies further confirm the effectiveness of our design.
Multi-Grained Multimodal Interaction Network for Entity Linking
Jul 19, 2023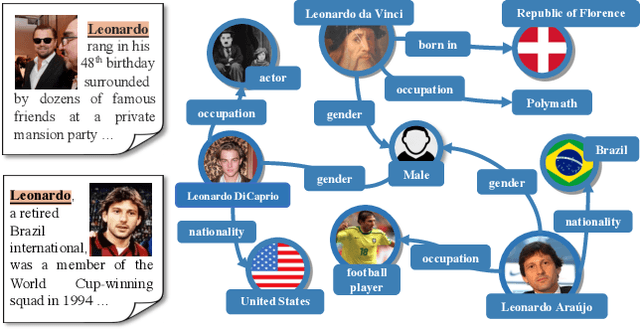
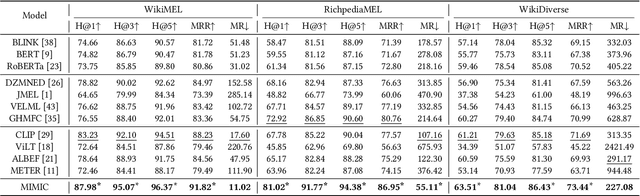


Abstract:Multimodal entity linking (MEL) task, which aims at resolving ambiguous mentions to a multimodal knowledge graph, has attracted wide attention in recent years. Though large efforts have been made to explore the complementary effect among multiple modalities, however, they may fail to fully absorb the comprehensive expression of abbreviated textual context and implicit visual indication. Even worse, the inevitable noisy data may cause inconsistency of different modalities during the learning process, which severely degenerates the performance. To address the above issues, in this paper, we propose a novel Multi-GraIned Multimodal InteraCtion Network $\textbf{(MIMIC)}$ framework for solving the MEL task. Specifically, the unified inputs of mentions and entities are first encoded by textual/visual encoders separately, to extract global descriptive features and local detailed features. Then, to derive the similarity matching score for each mention-entity pair, we device three interaction units to comprehensively explore the intra-modal interaction and inter-modal fusion among features of entities and mentions. In particular, three modules, namely the Text-based Global-Local interaction Unit (TGLU), Vision-based DuaL interaction Unit (VDLU) and Cross-Modal Fusion-based interaction Unit (CMFU) are designed to capture and integrate the fine-grained representation lying in abbreviated text and implicit visual cues. Afterwards, we introduce a unit-consistency objective function via contrastive learning to avoid inconsistency and model degradation. Experimental results on three public benchmark datasets demonstrate that our solution outperforms various state-of-the-art baselines, and ablation studies verify the effectiveness of designed modules.
Inheritance-guided Hierarchical Assignment for Clinical Automatic Diagnosis
Jan 27, 2021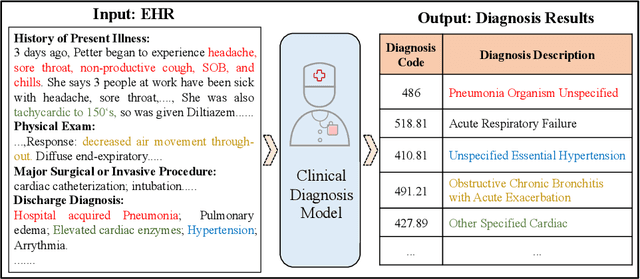
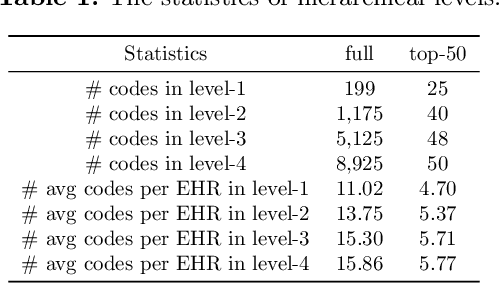
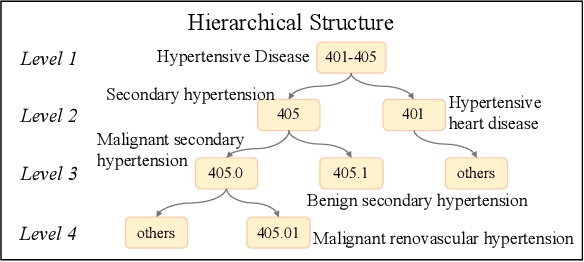
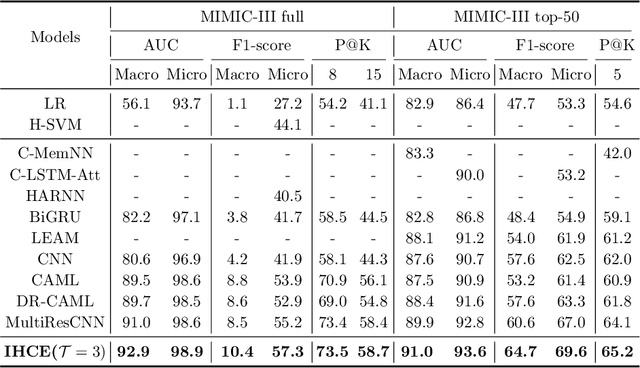
Abstract:Clinical diagnosis, which aims to assign diagnosis codes for a patient based on the clinical note, plays an essential role in clinical decision-making. Considering that manual diagnosis could be error-prone and time-consuming, many intelligent approaches based on clinical text mining have been proposed to perform automatic diagnosis. However, these methods may not achieve satisfactory results due to the following challenges. First, most of the diagnosis codes are rare, and the distribution is extremely unbalanced. Second, existing methods are challenging to capture the correlation between diagnosis codes. Third, the lengthy clinical note leads to the excessive dispersion of key information related to codes. To tackle these challenges, we propose a novel framework to combine the inheritance-guided hierarchical assignment and co-occurrence graph propagation for clinical automatic diagnosis. Specifically, we propose a hierarchical joint prediction strategy to address the challenge of unbalanced codes distribution. Then, we utilize graph convolutional neural networks to obtain the correlation and semantic representations of medical ontology. Furthermore, we introduce multi attention mechanisms to extract crucial information. Finally, extensive experiments on MIMIC-III dataset clearly validate the effectiveness of our method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge