Parth Patel
Domain Expansion: Parameter-Efficient Modules as Building Blocks for Composite Domains
Jan 24, 2025Abstract:Parameter-Efficient Fine-Tuning (PEFT) is an efficient alternative to full scale fine-tuning, gaining popularity recently. With pre-trained model sizes growing exponentially, PEFT can be effectively utilized to fine-tune compact modules, Parameter-Efficient Modules (PEMs), trained to be domain experts over diverse domains. In this project, we explore composing such individually fine-tuned PEMs for distribution generalization over the composite domain. To compose PEMs, simple composing functions are used that operate purely on the weight space of the individually fine-tuned PEMs, without requiring any additional fine-tuning. The proposed method is applied to the task of representing the 16 Myers-Briggs Type Indicator (MBTI) composite personalities via 4 building block dichotomies, comprising of 8 individual traits which can be merged (composed) to yield a unique personality. We evaluate the individual trait PEMs and the composed personality PEMs via an online MBTI personality quiz questionnaire, validating the efficacy of PEFT to fine-tune PEMs and merging PEMs without further fine-tuning for domain composition.
NeuraLunaDTNet: Feedforward Neural Network-Based Routing Protocol for Delay-Tolerant Lunar Communication Networks
Apr 07, 2024Abstract:Space Communication poses challenges such as severe delays, hard-to-predict routes and communication disruptions. The Delay Tolerant Network architecture, having been specifically designed keeping such scenarios in mind, is suitable to address some challenges. The traditional DTN routing protocols fall short of delivering optimal performance, due to the inherent complexities of space communication. Researchers have aimed at using recent advancements in AI to mitigate some routing challenges [9]. We propose utilising a feedforward neural network to develop a novel protocol NeuraLunaDTNet, which enhances the efficiency of the PRoPHET routing protocol for lunar communication, by learning contact plans in dynamically changing spatio-temporal graph.
APACE: AlphaFold2 and advanced computing as a service for accelerated discovery in biophysics
Aug 15, 2023Abstract:The prediction of protein 3D structure from amino acid sequence is a computational grand challenge in biophysics, and plays a key role in robust protein structure prediction algorithms, from drug discovery to genome interpretation. The advent of AI models, such as AlphaFold, is revolutionizing applications that depend on robust protein structure prediction algorithms. To maximize the impact, and ease the usability, of these novel AI tools we introduce APACE, AlphaFold2 and advanced computing as a service, a novel computational framework that effectively handles this AI model and its TB-size database to conduct accelerated protein structure prediction analyses in modern supercomputing environments. We deployed APACE in the Delta supercomputer, and quantified its performance for accurate protein structure predictions using four exemplar proteins: 6AWO, 6OAN, 7MEZ, and 6D6U. Using up to 200 ensembles, distributed across 50 nodes in Delta, equivalent to 200 A100 NVIDIA GPUs, we found that APACE is up to two orders of magnitude faster than off-the-shelf AlphaFold2 implementations, reducing time-to-solution from weeks to minutes. This computational approach may be readily linked with robotics laboratories to automate and accelerate scientific discovery.
Impossibility of Depth Reduction in Explainable Clustering
May 04, 2023Abstract:Over the last few years Explainable Clustering has gathered a lot of attention. Dasgupta et al. [ICML'20] initiated the study of explainable k-means and k-median clustering problems where the explanation is captured by a threshold decision tree which partitions the space at each node using axis parallel hyperplanes. Recently, Laber et al. [Pattern Recognition'23] made a case to consider the depth of the decision tree as an additional complexity measure of interest. In this work, we prove that even when the input points are in the Euclidean plane, then any depth reduction in the explanation incurs unbounded loss in the k-means and k-median cost. Formally, we show that there exists a data set X in the Euclidean plane, for which there is a decision tree of depth k-1 whose k-means/k-median cost matches the optimal clustering cost of X, but every decision tree of depth less than k-1 has unbounded cost w.r.t. the optimal cost of clustering. We extend our results to the k-center objective as well, albeit with weaker guarantees.
LT-GAN: Self-Supervised GAN with Latent Transformation Detection
Oct 19, 2020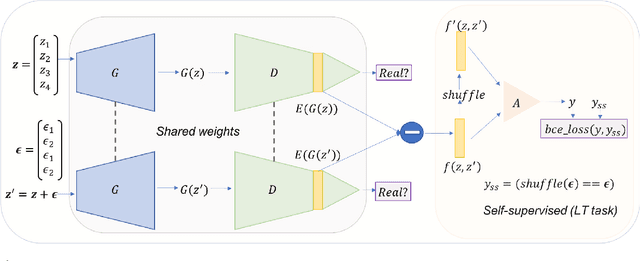
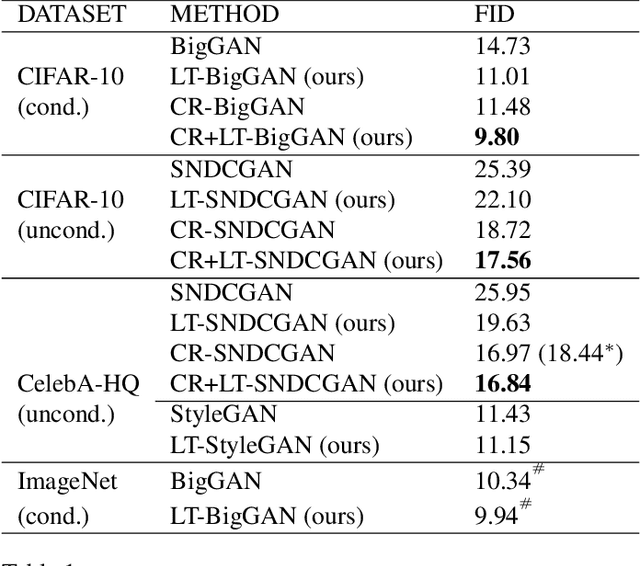
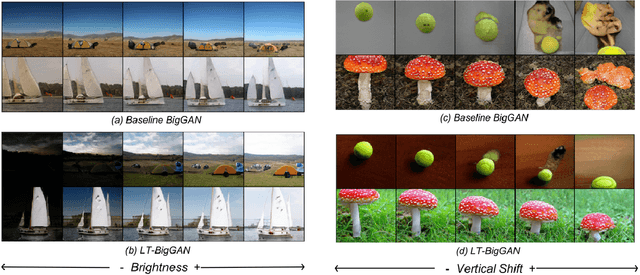
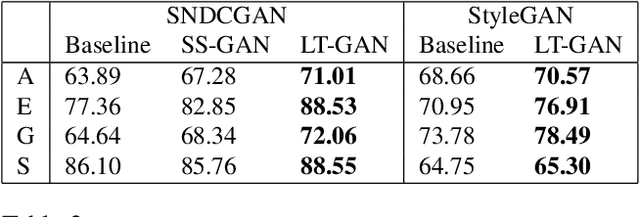
Abstract:Generative Adversarial Networks (GANs) coupled with self-supervised tasks have shown promising results in unconditional and semi-supervised image generation. We propose a self-supervised approach (LT-GAN) to improve the generation quality and diversity of images by estimating the GAN-induced transformation (i.e. transformation induced in the generated images by perturbing the latent space of generator). Specifically, given two pairs of images where each pair comprises of a generated image and its transformed version, the self-supervision task aims to identify whether the latent transformation applied in the given pair is same to that of the other pair. Hence, this auxiliary loss encourages the generator to produce images that are distinguishable by the auxiliary network, which in turn promotes the synthesis of semantically consistent images with respect to latent transformations. We show the efficacy of this pretext task by improving the image generation quality in terms of FID on state-of-the-art models for both conditional and unconditional settings on CIFAR-10, CelebA-HQ and ImageNet datasets. Moreover, we empirically show that LT-GAN helps in improving controlled image editing for CelebA-HQ and ImageNet over baseline models. We experimentally demonstrate that our proposed LT self-supervision task can be effectively combined with other state-of-the-art training techniques for added benefits. Consequently, we show that our approach achieves the new state-of-the-art FID score of 9.8 on conditional CIFAR-10 image generation.
Prediction of Cancer Microarray and DNA Methylation Data using Non-negative Matrix Factorization
Jul 15, 2020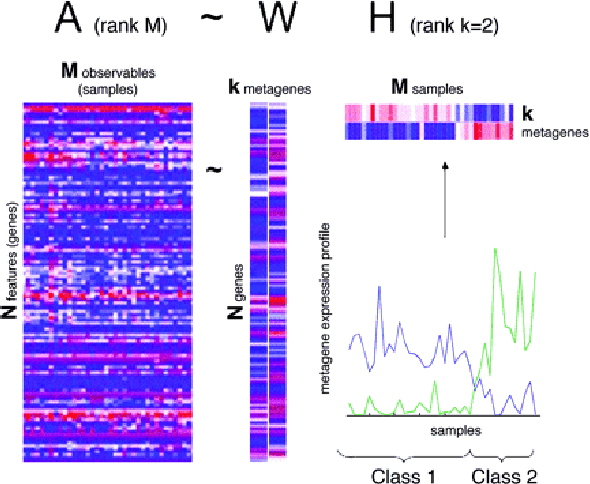
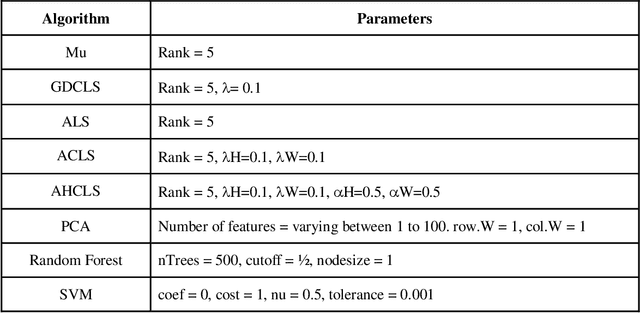
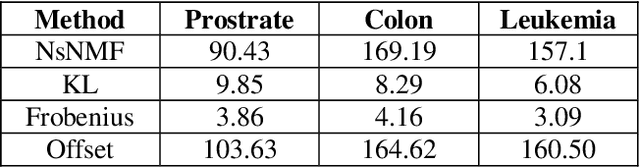
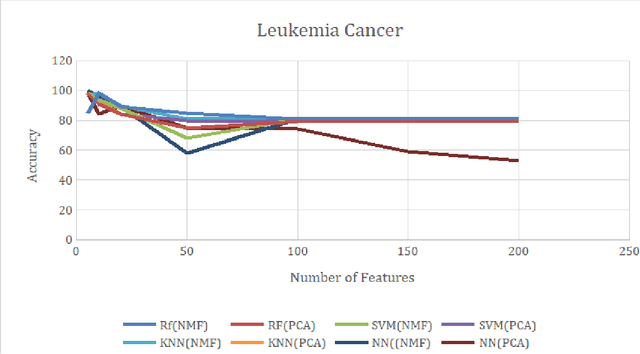
Abstract:Over the past few years, there has been a considerable spread of microarray technology in many biological patterns, particularly in those pertaining to cancer diseases like leukemia, prostate, colon cancer, etc. The primary bottleneck that one experiences in the proper understanding of such datasets lies in their dimensionality, and thus for an efficient and effective means of studying the same, a reduction in their dimension to a large extent is deemed necessary. This study is a bid to suggesting different algorithms and approaches for the reduction of dimensionality of such microarray datasets. This study exploits the matrix-like structure of such microarray data and uses a popular technique called Non-Negative Matrix Factorization (NMF) to reduce the dimensionality, primarily in the field of biological data. Classification accuracies are then compared for these algorithms. This technique gives an accuracy of 98%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge