Kalpdrum Passi
Prediction of Cancer Microarray and DNA Methylation Data using Non-negative Matrix Factorization
Jul 15, 2020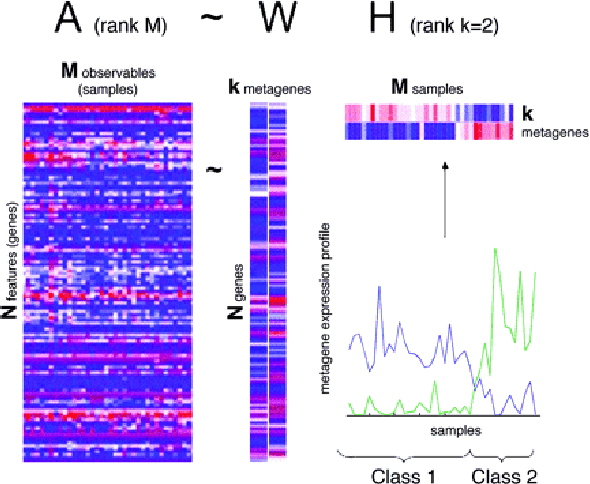
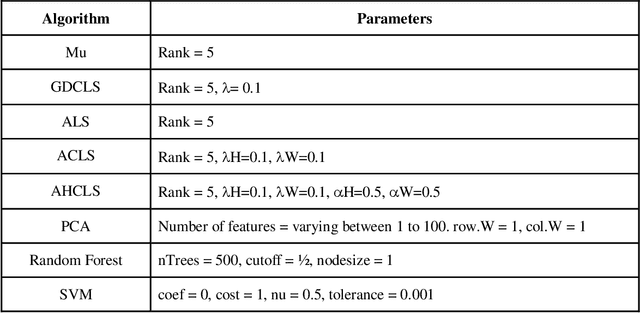
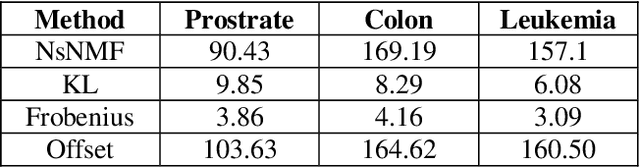
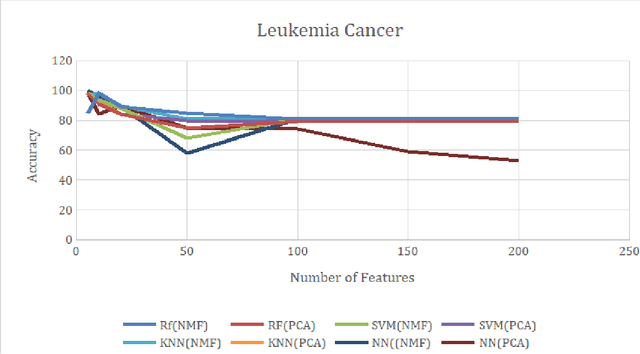
Abstract:Over the past few years, there has been a considerable spread of microarray technology in many biological patterns, particularly in those pertaining to cancer diseases like leukemia, prostate, colon cancer, etc. The primary bottleneck that one experiences in the proper understanding of such datasets lies in their dimensionality, and thus for an efficient and effective means of studying the same, a reduction in their dimension to a large extent is deemed necessary. This study is a bid to suggesting different algorithms and approaches for the reduction of dimensionality of such microarray datasets. This study exploits the matrix-like structure of such microarray data and uses a popular technique called Non-Negative Matrix Factorization (NMF) to reduce the dimensionality, primarily in the field of biological data. Classification accuracies are then compared for these algorithms. This technique gives an accuracy of 98%.
Increased Prediction Accuracy in the Game of Cricket using Machine Learning
Apr 09, 2018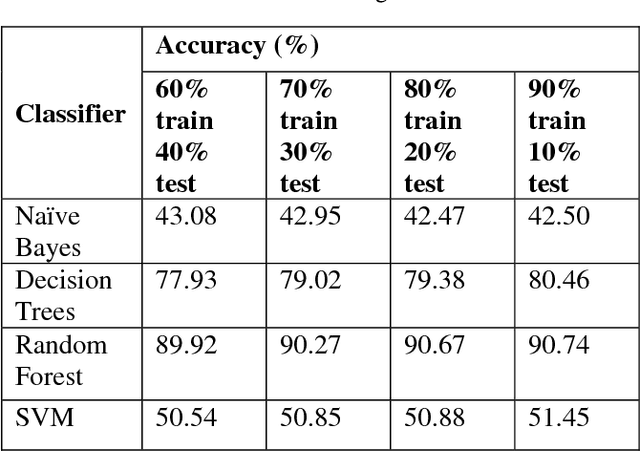
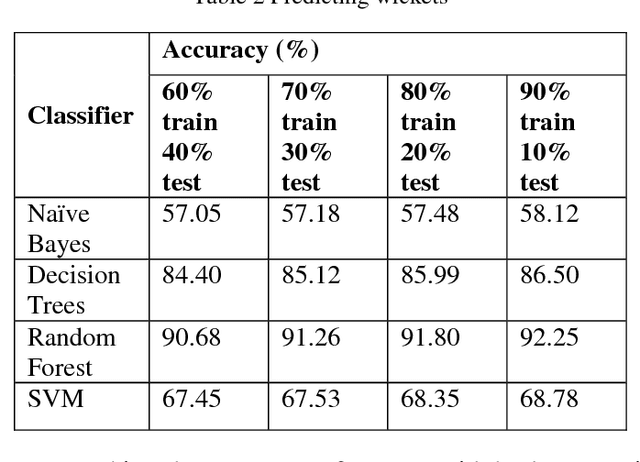
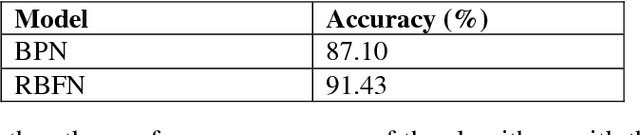
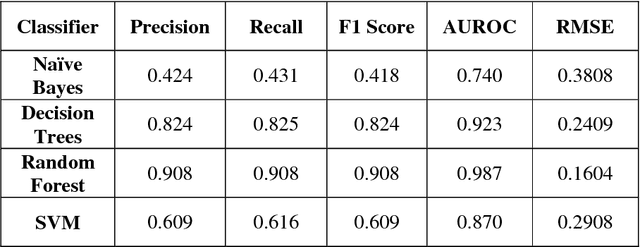
Abstract:Player selection is one the most important tasks for any sport and cricket is no exception. The performance of the players depends on various factors such as the opposition team, the venue, his current form etc. The team management, the coach and the captain select 11 players for each match from a squad of 15 to 20 players. They analyze different characteristics and the statistics of the players to select the best playing 11 for each match. Each batsman contributes by scoring maximum runs possible and each bowler contributes by taking maximum wickets and conceding minimum runs. This paper attempts to predict the performance of players as how many runs will each batsman score and how many wickets will each bowler take for both the teams. Both the problems are targeted as classification problems where number of runs and number of wickets are classified in different ranges. We used na\"ive bayes, random forest, multiclass SVM and decision tree classifiers to generate the prediction models for both the problems. Random Forest classifier was found to be the most accurate for both the problems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge