Pao-Hsiung Chiu
Out-of-Distribution Generalization for Neural Physics Solvers
Jan 27, 2026Abstract:Neural physics solvers are increasingly used in scientific discovery, given their potential for rapid in silico insights into physical, materials, or biological systems and their long-time evolution. However, poor generalization beyond their training support limits exploration of novel designs and long-time horizon predictions. We introduce NOVA, a route to generalizable neural physics solvers that can provide rapid, accurate solutions to scenarios even under distributional shifts in partial differential equation parameters, geometries and initial conditions. By learning physics-aligned representations from an initial sparse set of scenarios, NOVA consistently achieves 1-2 orders of magnitude lower out-of-distribution errors than data-driven baselines across complex, nonlinear problems including heat transfer, diffusion-reaction and fluid flow. We further showcase NOVA's dual impact on stabilizing long-time dynamical rollouts and improving generative design through application to the simulation of nonlinear Turing systems and fluidic chip optimization. Unlike neural physics solvers that are constrained to retrieval and/or emulation within an a priori space, NOVA enables reliable extrapolation beyond known regimes, a key capability given the need for exploration of novel hypothesis spaces in scientific discovery
Physics-Informed Uncertainty Enables Reliable AI-driven Design
Jan 26, 2026Abstract:Inverse design is a central goal in much of science and engineering, including frequency-selective surfaces (FSS) that are critical to microelectronics for telecommunications and optical metamaterials. Traditional surrogate-assisted optimization methods using deep learning can accelerate the design process but do not usually incorporate uncertainty quantification, leading to poorer optimization performance due to erroneous predictions in data-sparse regions. Here, we introduce and validate a fundamentally different paradigm of Physics-Informed Uncertainty, where the degree to which a model's prediction violates fundamental physical laws serves as a computationally-cheap and effective proxy for predictive uncertainty. By integrating physics-informed uncertainty into a multi-fidelity uncertainty-aware optimization workflow to design complex frequency-selective surfaces within the 20 - 30 GHz range, we increase the success rate of finding performant solutions from less than 10% to over 50%, while simultaneously reducing computational cost by an order of magnitude compared to the sole use of a high-fidelity solver. These results highlight the necessity of incorporating uncertainty quantification in machine-learning-driven inverse design for high-dimensional problems, and establish physics-informed uncertainty as a viable alternative to quantifying uncertainty in surrogate models for physical systems, thereby setting the stage for autonomous scientific discovery systems that can efficiently and robustly explore and evaluate candidate designs.
Multi-level datasets training method in Physics-Informed Neural Networks
Apr 30, 2025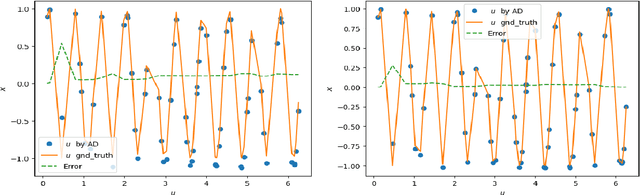
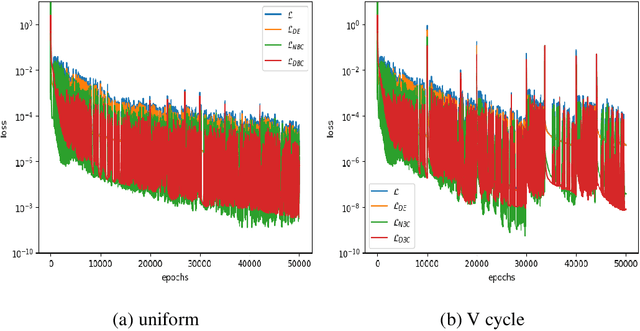
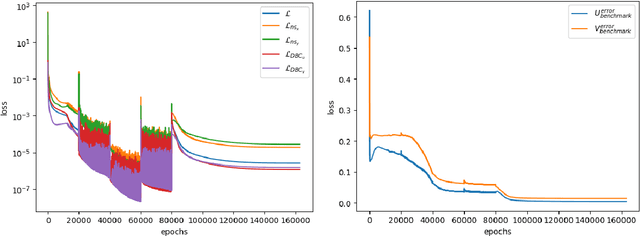
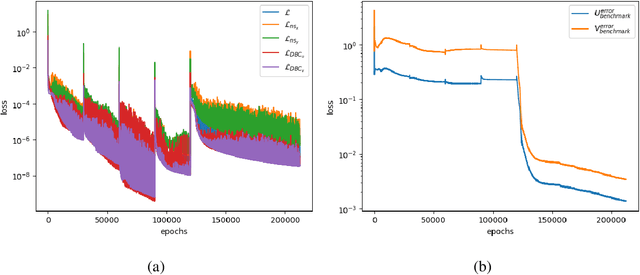
Abstract:Physics-Informed Neural Networks have emerged as a promising methodology for solving PDEs, gaining significant attention in computer science and various physics-related fields. Despite being demonstrated the ability to incorporate the physics of laws for versatile applications, PINNs still struggle with the challenging problems which are stiff to be solved and/or have high-frequency components in the solutions, resulting in accuracy and convergence issues. It may not only increase computational costs, but also lead to accuracy loss or solution divergence. In this study, an alternative approach is proposed to mitigate the above-mentioned problems. Inspired by the multi-grid method in CFD community, the underlying idea of the current approach is to efficiently remove different frequency errors via training with different levels of training samples, resulting in a simpler way to improve the training accuracy without spending time in fine-tuning of neural network structures, loss weights as well as hyperparameters. To demonstrate the efficacy of current approach, we first investigate canonical 1D ODE with high-frequency component and 2D convection-diffusion equation with V-cycle training strategy. Finally, the current method is employed for the classical benchmark problem of steady Lid-driven cavity flows at different Reynolds numbers, to investigate the applicability and efficacy for the problem involved multiple modes of high and low frequency. By virtue of various training sequence modes, improvement through predictions lead to 30% to 60% accuracy improvement. We also investigate the synergies between current method and transfer learning techniques for more challenging problems (i.e., higher Re). From the present results, it also revealed that the current framework can produce good predictions even for the case of Re=5000, demonstrating the ability to solve complex high-frequency PDEs.
Physics-Informed Neuro-Evolution (PINE): A Survey and Prospects
Jan 11, 2025Abstract:Deep learning models trained on finite data lack a complete understanding of the physical world. On the other hand, physics-informed neural networks (PINNs) are infused with such knowledge through the incorporation of mathematically expressible laws of nature into their training loss function. By complying with physical laws, PINNs provide advantages over purely data-driven models in limited-data regimes. This feature has propelled them to the forefront of scientific machine learning, a domain characterized by scarce and costly data. However, the vision of accurate physics-informed learning comes with significant challenges. This review examines PINNs for the first time in terms of model optimization and generalization, shedding light on the need for new algorithmic advances to overcome issues pertaining to the training speed, precision, and generalizability of today's PINN models. Of particular interest are the gradient-free methods of neuroevolution for optimizing the uniquely complex loss landscapes arising in PINN training. Methods synergizing gradient descent and neuroevolution for discovering bespoke neural architectures and balancing multiple conflicting terms in physics-informed learning objectives are positioned as important avenues for future research. Yet another exciting track is to cast neuroevolution as a meta-learner of generalizable PINN models.
Generalizable Neural Physics Solvers by Baldwinian Evolution
Dec 06, 2023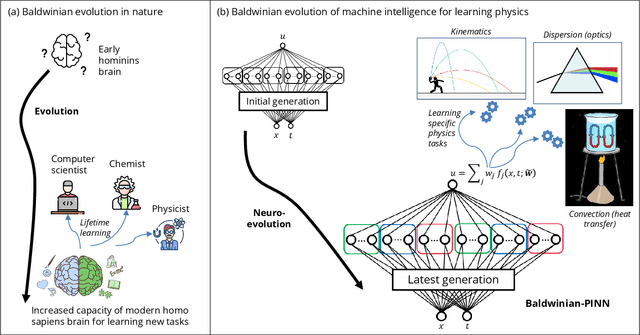
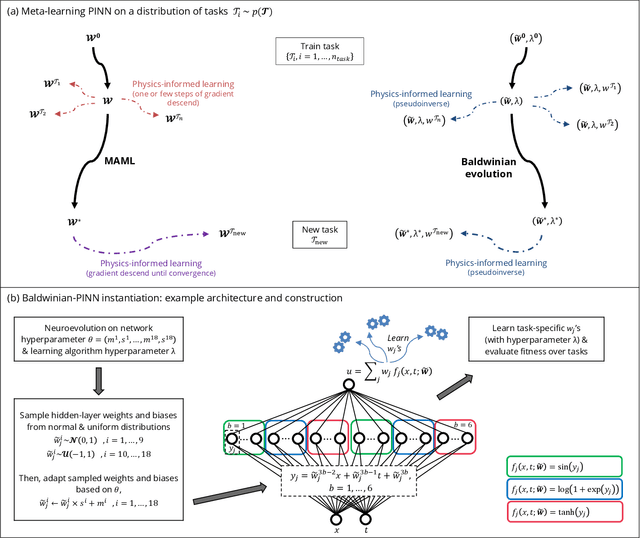
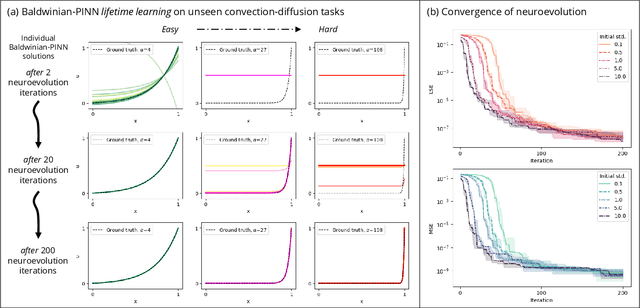

Abstract:Physics-informed neural networks (PINNs) are at the forefront of scientific machine learning, making possible the creation of machine intelligence that is cognizant of physical laws and able to accurately simulate them. In this paper, the potential of discovering PINNs that generalize over an entire family of physics tasks is studied, for the first time, through a biological lens of the Baldwin effect. Drawing inspiration from the neurodevelopment of precocial species that have evolved to learn, predict and react quickly to their environment, we envision PINNs that are pre-wired with connection strengths inducing strong biases towards efficient learning of physics. To this end, evolutionary selection pressure (guided by proficiency over a family of tasks) is coupled with lifetime learning (to specialize on a smaller subset of those tasks) to produce PINNs that demonstrate fast and physics-compliant prediction capabilities across a range of empirically challenging problem instances. The Baldwinian approach achieves an order of magnitude improvement in prediction accuracy at a fraction of the computation cost compared to state-of-the-art results with PINNs meta-learned by gradient descent. This paper marks a leap forward in the meta-learning of PINNs as generalizable physics solvers.
LSA-PINN: Linear Boundary Connectivity Loss for Solving PDEs on Complex Geometry
Feb 03, 2023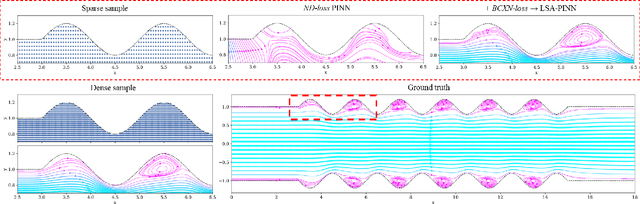
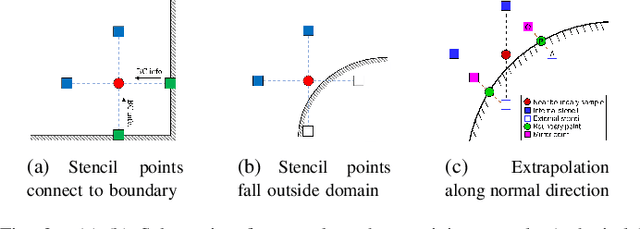
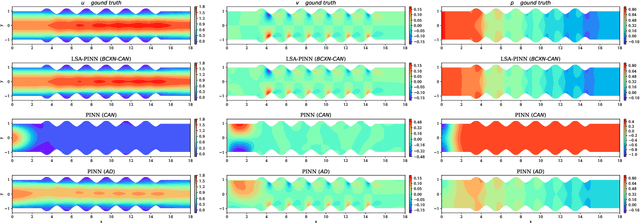
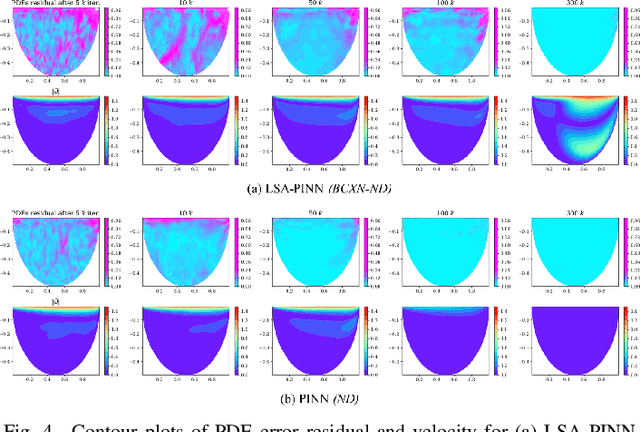
Abstract:We present a novel loss formulation for efficient learning of complex dynamics from governing physics, typically described by partial differential equations (PDEs), using physics-informed neural networks (PINNs). In our experiments, existing versions of PINNs are seen to learn poorly in many problems, especially for complex geometries, as it becomes increasingly difficult to establish appropriate sampling strategy at the near boundary region. Overly dense sampling can adversely impede training convergence if the local gradient behaviors are too complex to be adequately modelled by PINNs. On the other hand, if the samples are too sparse, existing PINNs tend to overfit the near boundary region, leading to incorrect solution. To prevent such issues, we propose a new Boundary Connectivity (BCXN) loss function which provides linear local structure approximation (LSA) to the gradient behaviors at the boundary for PINN. Our BCXN-loss implicitly imposes local structure during training, thus facilitating fast physics-informed learning across entire problem domains with order of magnitude sparser training samples. This LSA-PINN method shows a few orders of magnitude smaller errors than existing methods in terms of the standard L2-norm metric, while using dramatically fewer training samples and iterations. Our proposed LSA-PINN does not pose any requirement on the differentiable property of the networks, and we demonstrate its benefits and ease of implementation on both multi-layer perceptron and convolutional neural network versions as commonly used in current PINN literature.
JAX-Accelerated Neuroevolution of Physics-informed Neural Networks: Benchmarks and Experimental Results
Dec 15, 2022



Abstract:This paper introduces the use of evolutionary algorithms for solving differential equations. The solution is obtained by optimizing a deep neural network whose loss function is defined by the residual terms from the differential equations. Recent studies have used stochastic gradient descent (SGD) variants to train these physics-informed neural networks (PINNs), but these methods can struggle to find accurate solutions due to optimization challenges. When solving differential equations, it is important to find the globally optimum parameters of the network, rather than just finding a solution that works well during training. SGD only searches along a single gradient direction, so it may not be the best approach for training PINNs with their accompanying complex optimization landscapes. In contrast, evolutionary algorithms perform a parallel exploration of different solutions in order to avoid getting stuck in local optima and can potentially find more accurate solutions. However, evolutionary algorithms can be slow, which can make them difficult to use in practice. To address this, we provide a set of five benchmark problems with associated performance metrics and baseline results to support the development of evolutionary algorithms for enhanced PINN training. As a baseline, we evaluate the performance and speed of using the widely adopted Covariance Matrix Adaptation Evolution Strategy (CMA-ES) for solving PINNs. We provide the loss and training time for CMA-ES run on TensorFlow, and CMA-ES and SGD run on JAX (with GPU acceleration) for the five benchmark problems. Our results show that JAX-accelerated evolutionary algorithms, particularly CMA-ES, can be a useful approach for solving differential equations. We hope that our work will support the exploration and development of alternative optimization algorithms for the complex task of optimizing PINNs.
Design of Turing Systems with Physics-Informed Neural Networks
Nov 24, 2022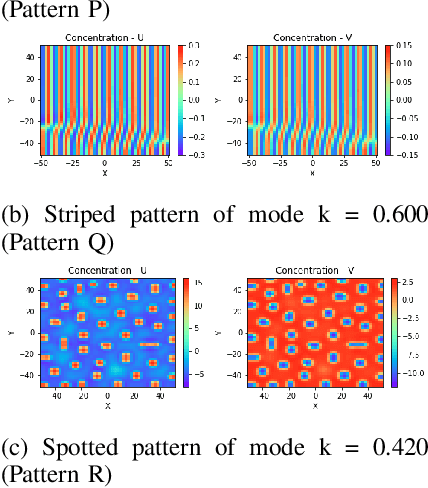
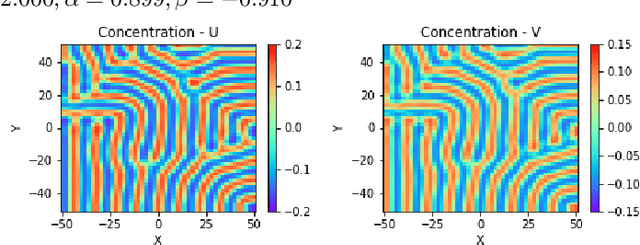
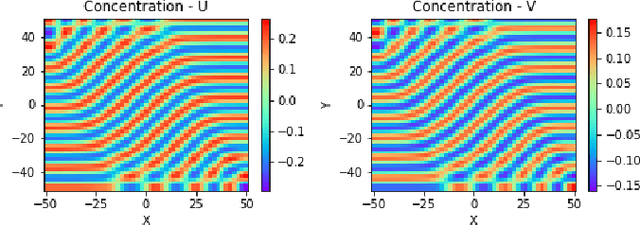
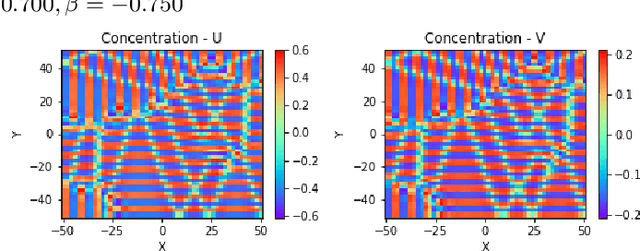
Abstract:Reaction-diffusion (Turing) systems are fundamental to the formation of spatial patterns in nature and engineering. These systems are governed by a set of non-linear partial differential equations containing parameters that determine the rate of constituent diffusion and reaction. Critically, these parameters, such as diffusion coefficient, heavily influence the mode and type of the final pattern, and quantitative characterization and knowledge of these parameters can aid in bio-mimetic design or understanding of real-world systems. However, the use of numerical methods to infer these parameters can be difficult and computationally expensive. Typically, adjoint solvers may be used, but they are frequently unstable for very non-linear systems. Alternatively, massive amounts of iterative forward simulations are used to find the best match, but this is extremely effortful. Recently, physics-informed neural networks have been proposed as a means for data-driven discovery of partial differential equations, and have seen success in various applications. Thus, we investigate the use of physics-informed neural networks as a tool to infer key parameters in reaction-diffusion systems in the steady-state for scientific discovery or design. Our proof-of-concept results show that the method is able to infer parameters for different pattern modes and types with errors of less than 10\%. In addition, the stochastic nature of this method can be exploited to provide multiple parameter alternatives to the desired pattern, highlighting the versatility of this method for bio-mimetic design. This work thus demonstrates the utility of physics-informed neural networks for inverse parameter inference of reaction-diffusion systems to enhance scientific discovery and design.
Robustness of Physics-Informed Neural Networks to Noise in Sensor Data
Nov 22, 2022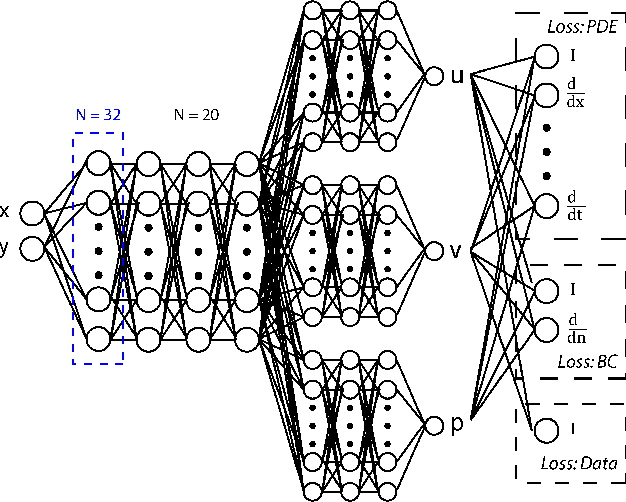
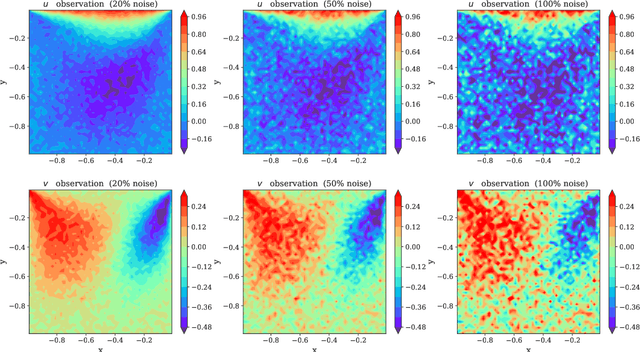
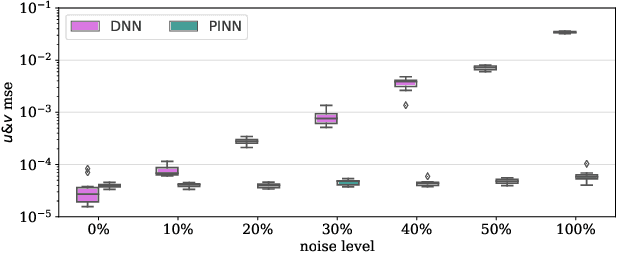
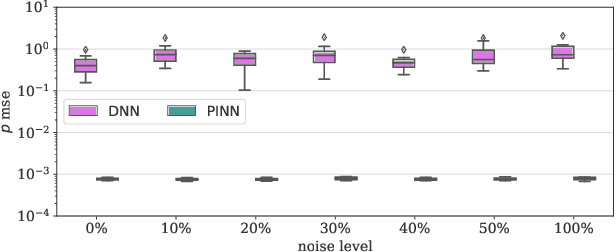
Abstract:Physics-Informed Neural Networks (PINNs) have been shown to be an effective way of incorporating physics-based domain knowledge into neural network models for many important real-world systems. They have been particularly effective as a means of inferring system information based on data, even in cases where data is scarce. Most of the current work however assumes the availability of high-quality data. In this work, we further conduct a preliminary investigation of the robustness of physics-informed neural networks to the magnitude of noise in the data. Interestingly, our experiments reveal that the inclusion of physics in the neural network is sufficient to negate the impact of noise in data originating from hypothetical low quality sensors with high signal-to-noise ratios of up to 1. The resultant predictions for this test case are seen to still match the predictive value obtained for equivalent data obtained from high-quality sensors with potentially 10x less noise. This further implies the utility of physics-informed neural network modeling for making sense of data from sensor networks in the future, especially with the advent of Industry 4.0 and the increasing trend towards ubiquitous deployment of low-cost sensors which are typically noisier.
CAN-PINN: A Fast Physics-Informed Neural Network Based on Coupled-Automatic-Numerical Differentiation Method
Oct 29, 2021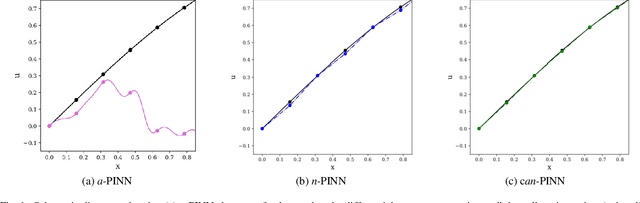
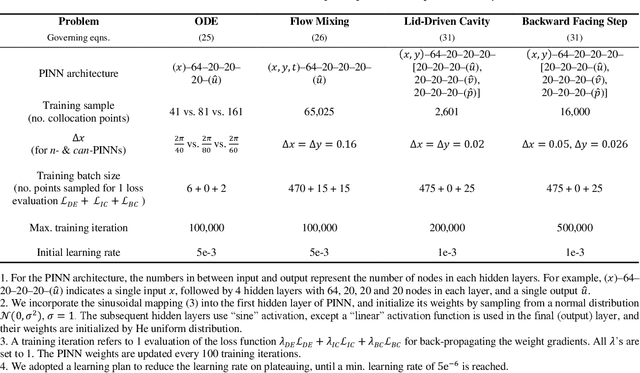
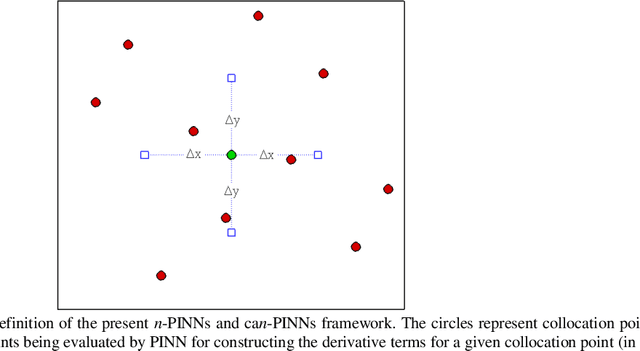
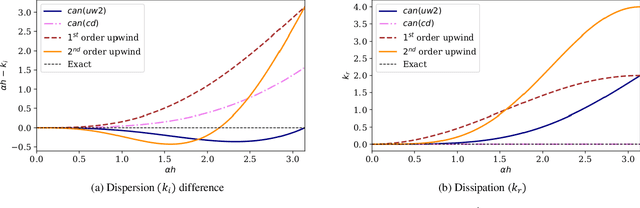
Abstract:In this study, novel physics-informed neural network (PINN) methods for coupling neighboring support points and automatic differentiation (AD) through Taylor series expansion are proposed to allow efficient training with improved accuracy. The computation of differential operators required for PINNs loss evaluation at collocation points are conventionally obtained via AD. Although AD has the advantage of being able to compute the exact gradients at any point, such PINNs can only achieve high accuracies with large numbers of collocation points, otherwise they are prone to optimizing towards unphysical solution. To make PINN training fast, the dual ideas of using numerical differentiation (ND)-inspired method and coupling it with AD are employed to define the loss function. The ND-based formulation for training loss can strongly link neighboring collocation points to enable efficient training in sparse sample regimes, but its accuracy is restricted by the interpolation scheme. The proposed coupled-automatic-numerical differentiation framework, labeled as can-PINN, unifies the advantages of AD and ND, providing more robust and efficient training than AD-based PINNs, while further improving accuracy by up to 1-2 orders of magnitude relative to ND-based PINNs. For a proof-of-concept demonstration of this can-scheme to fluid dynamic problems, two numerical-inspired instantiations of can-PINN schemes for the convection and pressure gradient terms were derived to solve the incompressible Navier-Stokes (N-S) equations. The superior performance of can-PINNs is demonstrated on several challenging problems, including the flow mixing phenomena, lid driven flow in a cavity, and channel flow over a backward facing step. The results reveal that for challenging problems like these, can-PINNs can consistently achieve very good accuracy whereas conventional AD-based PINNs fail.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge