Ozan Özdenizci
A Scalable Hybrid Training Approach for Recurrent Spiking Neural Networks
Jun 17, 2025Abstract:Recurrent spiking neural networks (RSNNs) can be implemented very efficiently in neuromorphic systems. Nevertheless, training of these models with powerful gradient-based learning algorithms is mostly performed on standard digital hardware using Backpropagation through time (BPTT). However, BPTT has substantial limitations. It does not permit online training and its memory consumption scales linearly with the number of computation steps. In contrast, learning methods using forward propagation of gradients operate in an online manner with a memory consumption independent of the number of time steps. These methods enable SNNs to learn from continuous, infinite-length input sequences. Yet, slow execution speed on conventional hardware as well as inferior performance has hindered their widespread application. In this work, we introduce HYbrid PRopagation (HYPR) that combines the efficiency of parallelization with approximate online forward learning. Our algorithm yields high-throughput online learning through parallelization, paired with constant, i.e., sequence length independent, memory demands. HYPR enables parallelization of parameter update computation over the sub sequences for RSNNs consisting of almost arbitrary non-linear spiking neuron models. We apply HYPR to networks of spiking neurons with oscillatory subthreshold dynamics. We find that this type of neuron model is particularly well trainable by HYPR, resulting in an unprecedentedly low task performance gap between approximate forward gradient learning and BPTT.
ViTaPEs: Visuotactile Position Encodings for Cross-Modal Alignment in Multimodal Transformers
May 26, 2025Abstract:Tactile sensing provides local essential information that is complementary to visual perception, such as texture, compliance, and force. Despite recent advances in visuotactile representation learning, challenges remain in fusing these modalities and generalizing across tasks and environments without heavy reliance on pre-trained vision-language models. Moreover, existing methods do not study positional encodings, thereby overlooking the multi-scale spatial reasoning needed to capture fine-grained visuotactile correlations. We introduce ViTaPEs, a transformer-based framework that robustly integrates visual and tactile input data to learn task-agnostic representations for visuotactile perception. Our approach exploits a novel multi-scale positional encoding scheme to capture intra-modal structures, while simultaneously modeling cross-modal cues. Unlike prior work, we provide provable guarantees in visuotactile fusion, showing that our encodings are injective, rigid-motion-equivariant, and information-preserving, validating these properties empirically. Experiments on multiple large-scale real-world datasets show that ViTaPEs not only surpasses state-of-the-art baselines across various recognition tasks but also demonstrates zero-shot generalization to unseen, out-of-domain scenarios. We further demonstrate the transfer-learning strength of ViTaPEs in a robotic grasping task, where it outperforms state-of-the-art baselines in predicting grasp success. Project page: https://sites.google.com/view/vitapes
Privacy-Aware Lifelong Learning
May 16, 2025Abstract:Lifelong learning algorithms enable models to incrementally acquire new knowledge without forgetting previously learned information. Contrarily, the field of machine unlearning focuses on explicitly forgetting certain previous knowledge from pretrained models when requested, in order to comply with data privacy regulations on the right-to-be-forgotten. Enabling efficient lifelong learning with the capability to selectively unlearn sensitive information from models presents a critical and largely unaddressed challenge with contradicting objectives. We address this problem from the perspective of simultaneously preventing catastrophic forgetting and allowing forward knowledge transfer during task-incremental learning, while ensuring exact task unlearning and minimizing memory requirements, based on a single neural network model to be adapted. Our proposed solution, privacy-aware lifelong learning (PALL), involves optimization of task-specific sparse subnetworks with parameter sharing within a single architecture. We additionally utilize an episodic memory rehearsal mechanism to facilitate exact unlearning without performance degradations. We empirically demonstrate the scalability of PALL across various architectures in image classification, and provide a state-of-the-art solution that uniquely integrates lifelong learning and privacy-aware unlearning mechanisms for responsible AI applications.
ReLI: A Language-Agnostic Approach to Human-Robot Interaction
May 03, 2025Abstract:Adapting autonomous agents to industrial, domestic, and other daily tasks is currently gaining momentum. However, in the global or cross-lingual application contexts, ensuring effective interaction with the environment and executing unrestricted human task-specified instructions in diverse languages remains an unsolved problem. To address this challenge, we propose ReLI, a language-agnostic framework designed to enable autonomous agents to converse naturally, semantically reason about the environment, and to perform downstream tasks, regardless of the task instruction's linguistic origin. First, we ground large-scale pre-trained foundation models and transform them into language-to-action models that can directly provide common-sense reasoning and high-level robot control through natural, free-flow human-robot conversational interactions. Further, we perform cross-lingual grounding of the models to ensure that ReLI generalises across the global languages. To demonstrate the ReLI's robustness, we conducted extensive simulated and real-world experiments on various short- and long-horizon tasks, including zero-shot and few-shot spatial navigation, scene information retrieval, and query-oriented tasks. We benchmarked the performance on 140 languages involving over 70K multi-turn conversations. On average, ReLI achieved over 90%$\pm$0.2 accuracy in cross-lingual instruction parsing and task execution success rates. These results demonstrate the ReLI's potential to enhance natural human-robot interaction in the real world while championing linguistic diversity. Demonstrations and resources will be publicly available at https://linusnep.github.io/ReLI/.
Adversarially Robust Spiking Neural Networks Through Conversion
Nov 15, 2023Abstract:Spiking neural networks (SNNs) provide an energy-efficient alternative to a variety of artificial neural network (ANN) based AI applications. As the progress in neuromorphic computing with SNNs expands their use in applications, the problem of adversarial robustness of SNNs becomes more pronounced. To the contrary of the widely explored end-to-end adversarial training based solutions, we address the limited progress in scalable robust SNN training methods by proposing an adversarially robust ANN-to-SNN conversion algorithm. Our method provides an efficient approach to embrace various computationally demanding robust learning objectives that have been proposed for ANNs. During a post-conversion robust finetuning phase, our method adversarially optimizes both layer-wise firing thresholds and synaptic connectivity weights of the SNN to maintain transferred robustness gains from the pre-trained ANN. We perform experimental evaluations in numerous adaptive adversarial settings that account for the spike-based operation dynamics of SNNs, and show that our approach yields a scalable state-of-the-art solution for adversarially robust deep SNNs with low-latency.
TS-MoCo: Time-Series Momentum Contrast for Self-Supervised Physiological Representation Learning
Jun 10, 2023Abstract:Limited availability of labeled physiological data often prohibits the use of powerful supervised deep learning models in the biomedical machine intelligence domain. We approach this problem and propose a novel encoding framework that relies on self-supervised learning with momentum contrast to learn representations from multivariate time-series of various physiological domains without needing labels. Our model uses a transformer architecture that can be easily adapted to classification problems by optimizing a linear output classification layer. We experimentally evaluate our framework using two publicly available physiological datasets from different domains, i.e., human activity recognition from embedded inertial sensory and emotion recognition from electroencephalography. We show that our self-supervised learning approach can indeed learn discriminative features which can be exploited in downstream classification tasks. Our work enables the development of domain-agnostic intelligent systems that can effectively analyze multivariate time-series data from physiological domains.
Restoring Vision in Adverse Weather Conditions with Patch-Based Denoising Diffusion Models
Jul 29, 2022



Abstract:Image restoration under adverse weather conditions has been of significant interest for various computer vision applications. Recent successful methods rely on the current progress in deep neural network architectural designs (e.g., with vision transformers). Motivated by the recent progress achieved with state-of-the-art conditional generative models, we present a novel patch-based image restoration algorithm based on denoising diffusion probabilistic models. Our patch-based diffusion modeling approach enables size-agnostic image restoration by using a guided denoising process with smoothed noise estimates across overlapping patches during inference. We empirically evaluate our model on benchmark datasets for image desnowing, combined deraining and dehazing, and raindrop removal. We demonstrate our approach to achieve state-of-the-art performances on both weather-specific and multi-weather image restoration, and qualitatively show strong generalization to real-world test images.
EEG2Vec: Learning Affective EEG Representations via Variational Autoencoders
Jul 16, 2022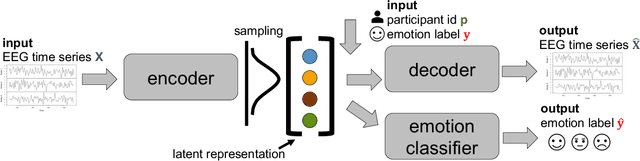
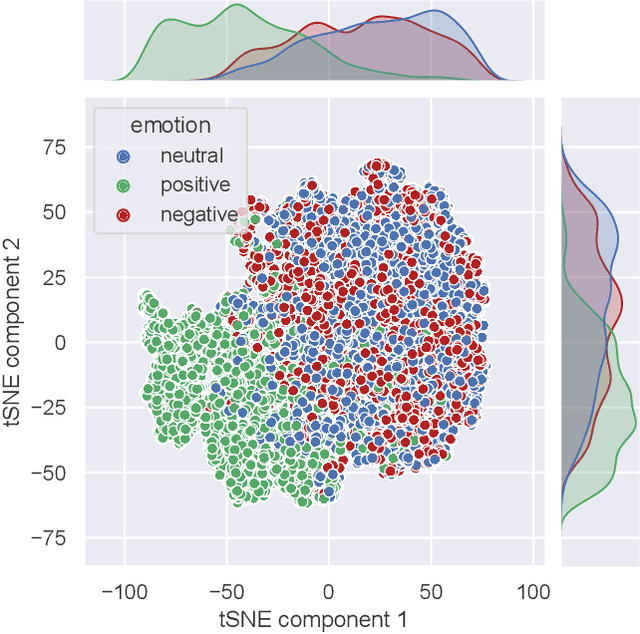
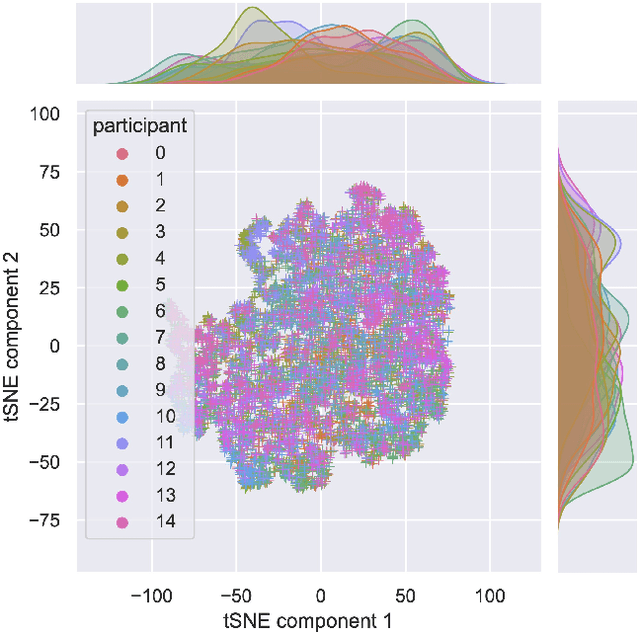
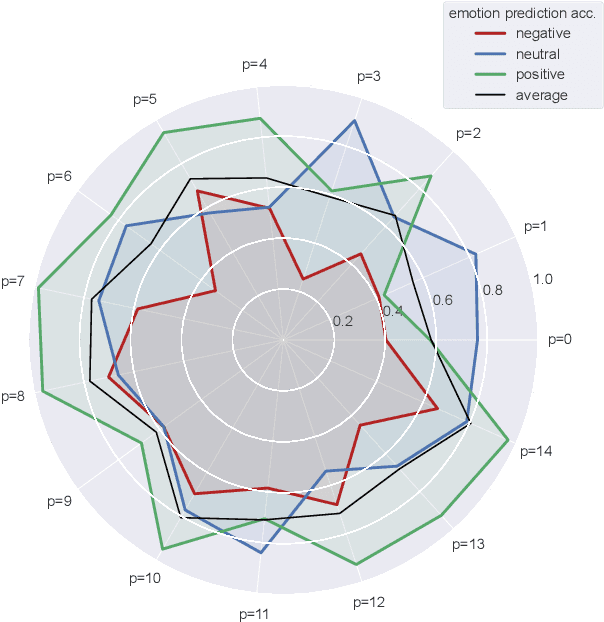
Abstract:There is a growing need for sparse representational formats of human affective states that can be utilized in scenarios with limited computational memory resources. We explore whether representing neural data, in response to emotional stimuli, in a latent vector space can serve to both predict emotional states as well as generate synthetic EEG data that are participant- and/or emotion-specific. We propose a conditional variational autoencoder based framework, EEG2Vec, to learn generative-discriminative representations from EEG data. Experimental results on affective EEG recording datasets demonstrate that our model is suitable for unsupervised EEG modeling, classification of three distinct emotion categories (positive, neutral, negative) based on the latent representation achieves a robust performance of 68.49%, and generated synthetic EEG sequences resemble real EEG data inputs to particularly reconstruct low-frequency signal components. Our work advances areas where affective EEG representations can be useful in e.g., generating artificial (labeled) training data or alleviating manual feature extraction, and provide efficiency for memory constrained edge computing applications.
Memory-enriched computation and learning in spiking neural networks through Hebbian plasticity
May 23, 2022



Abstract:Memory is a key component of biological neural systems that enables the retention of information over a huge range of temporal scales, ranging from hundreds of milliseconds up to years. While Hebbian plasticity is believed to play a pivotal role in biological memory, it has so far been analyzed mostly in the context of pattern completion and unsupervised learning. Here, we propose that Hebbian plasticity is fundamental for computations in biological neural systems. We introduce a novel spiking neural network architecture that is enriched by Hebbian synaptic plasticity. We show that Hebbian enrichment renders spiking neural networks surprisingly versatile in terms of their computational as well as learning capabilities. It improves their abilities for out-of-distribution generalization, one-shot learning, cross-modal generative association, language processing, and reward-based learning. As spiking neural networks are the basis for energy-efficient neuromorphic hardware, this also suggests that powerful cognitive neuromorphic systems can be build based on this principle.
Exploiting Multiple EEG Data Domains with Adversarial Learning
Apr 16, 2022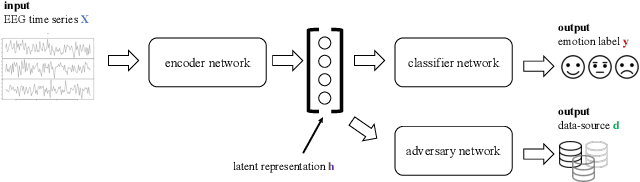
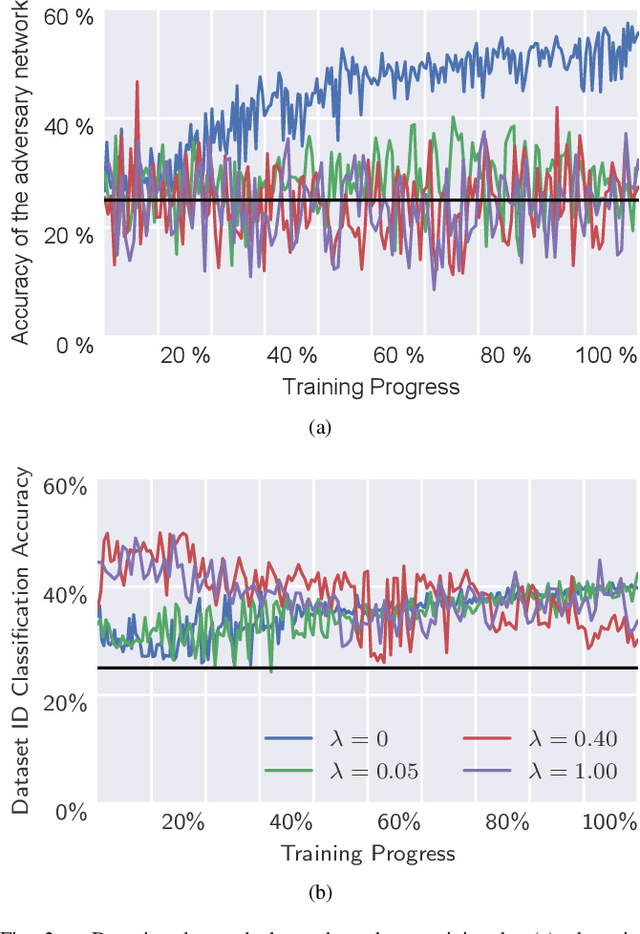
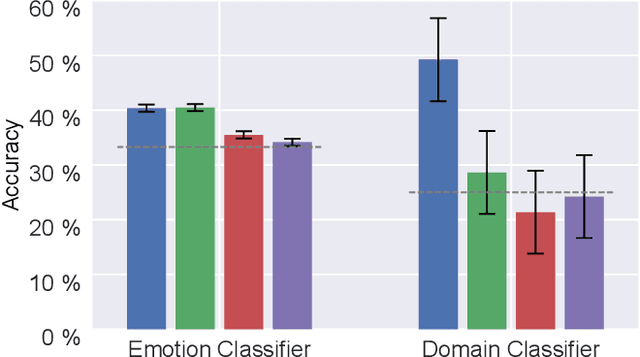
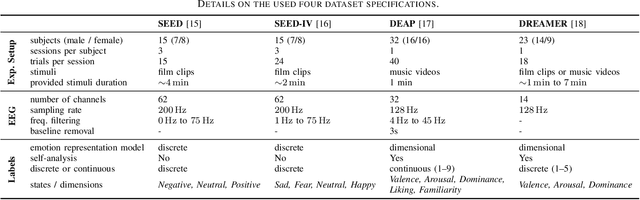
Abstract:Electroencephalography (EEG) is shown to be a valuable data source for evaluating subjects' mental states. However, the interpretation of multi-modal EEG signals is challenging, as they suffer from poor signal-to-noise-ratio, are highly subject-dependent, and are bound to the equipment and experimental setup used, (i.e. domain). This leads to machine learning models often suffer from poor generalization ability, where they perform significantly worse on real-world data than on the exploited training data. Recent research heavily focuses on cross-subject and cross-session transfer learning frameworks to reduce domain calibration efforts for EEG signals. We argue that multi-source learning via learning domain-invariant representations from multiple data-sources is a viable alternative, as the available data from different EEG data-source domains (e.g., subjects, sessions, experimental setups) grow massively. We propose an adversarial inference approach to learn data-source invariant representations in this context, enabling multi-source learning for EEG-based brain-computer interfaces. We unify EEG recordings from different source domains (i.e., emotion recognition datasets SEED, SEED-IV, DEAP, DREAMER), and demonstrate the feasibility of our invariant representation learning approach in suppressing data-source-relevant information leakage by 35% while still achieving stable EEG-based emotion classification performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge