Tobias Grosse-Puppendahl
HappyRouting: Learning Emotion-Aware Route Trajectories for Scalable In-The-Wild Navigation
Jan 28, 2024Abstract:Routes represent an integral part of triggering emotions in drivers. Navigation systems allow users to choose a navigation strategy, such as the fastest or shortest route. However, they do not consider the driver's emotional well-being. We present HappyRouting, a novel navigation-based empathic car interface guiding drivers through real-world traffic while evoking positive emotions. We propose design considerations, derive a technical architecture, and implement a routing optimization framework. Our contribution is a machine learning-based generated emotion map layer, predicting emotions along routes based on static and dynamic contextual data. We evaluated HappyRouting in a real-world driving study (N=13), finding that happy routes increase subjectively perceived valence by 11% (p=.007). Although happy routes take 1.25 times longer on average, participants perceived the happy route as shorter, presenting an emotion-enhanced alternative to today's fastest routing mechanisms. We discuss how emotion-based routing can be integrated into navigation apps, promoting emotional well-being for mobility use.
TS-MoCo: Time-Series Momentum Contrast for Self-Supervised Physiological Representation Learning
Jun 10, 2023Abstract:Limited availability of labeled physiological data often prohibits the use of powerful supervised deep learning models in the biomedical machine intelligence domain. We approach this problem and propose a novel encoding framework that relies on self-supervised learning with momentum contrast to learn representations from multivariate time-series of various physiological domains without needing labels. Our model uses a transformer architecture that can be easily adapted to classification problems by optimizing a linear output classification layer. We experimentally evaluate our framework using two publicly available physiological datasets from different domains, i.e., human activity recognition from embedded inertial sensory and emotion recognition from electroencephalography. We show that our self-supervised learning approach can indeed learn discriminative features which can be exploited in downstream classification tasks. Our work enables the development of domain-agnostic intelligent systems that can effectively analyze multivariate time-series data from physiological domains.
EEG2Vec: Learning Affective EEG Representations via Variational Autoencoders
Jul 16, 2022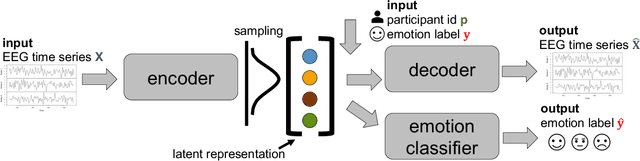
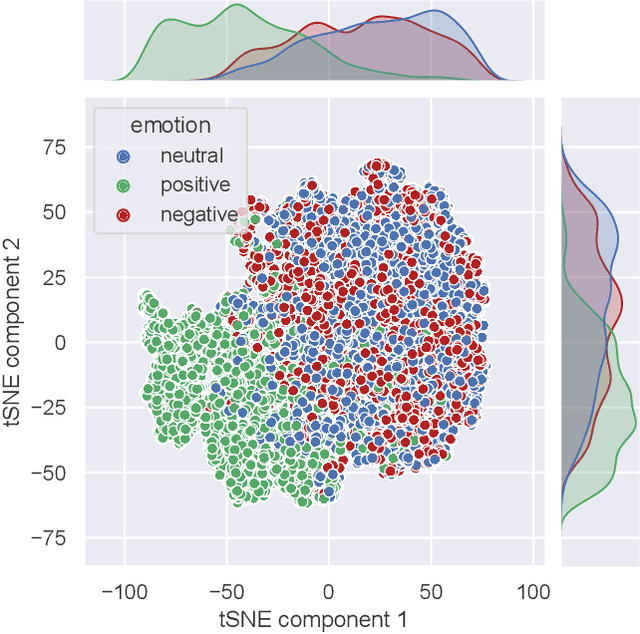
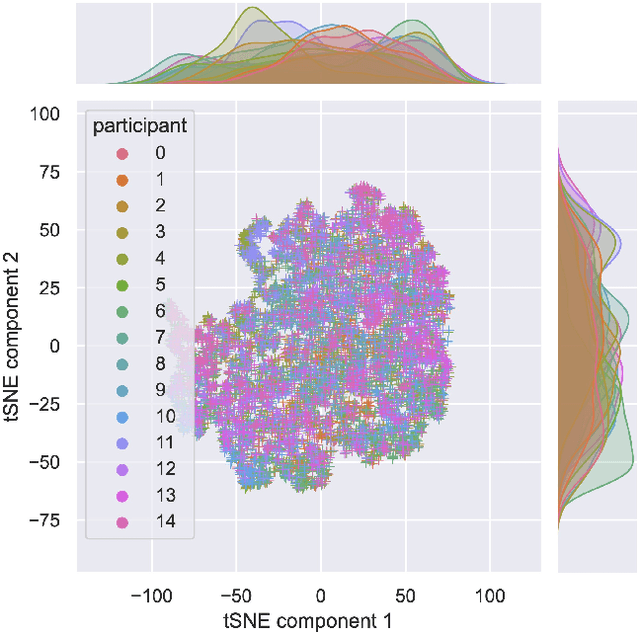
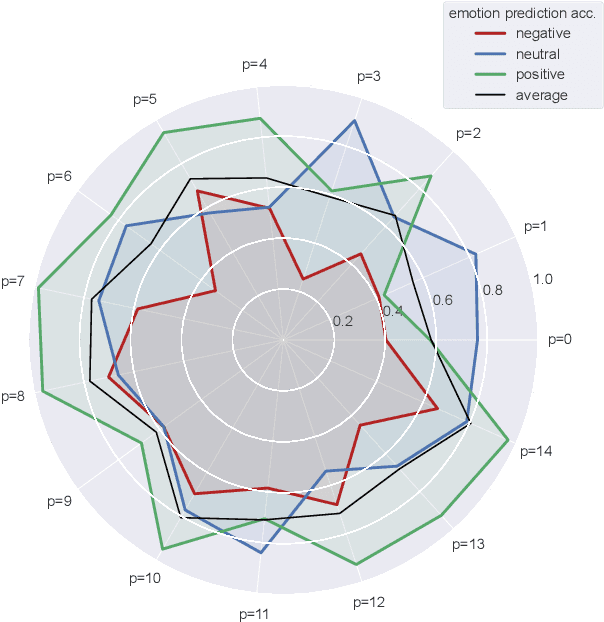
Abstract:There is a growing need for sparse representational formats of human affective states that can be utilized in scenarios with limited computational memory resources. We explore whether representing neural data, in response to emotional stimuli, in a latent vector space can serve to both predict emotional states as well as generate synthetic EEG data that are participant- and/or emotion-specific. We propose a conditional variational autoencoder based framework, EEG2Vec, to learn generative-discriminative representations from EEG data. Experimental results on affective EEG recording datasets demonstrate that our model is suitable for unsupervised EEG modeling, classification of three distinct emotion categories (positive, neutral, negative) based on the latent representation achieves a robust performance of 68.49%, and generated synthetic EEG sequences resemble real EEG data inputs to particularly reconstruct low-frequency signal components. Our work advances areas where affective EEG representations can be useful in e.g., generating artificial (labeled) training data or alleviating manual feature extraction, and provide efficiency for memory constrained edge computing applications.
Exploiting Multiple EEG Data Domains with Adversarial Learning
Apr 16, 2022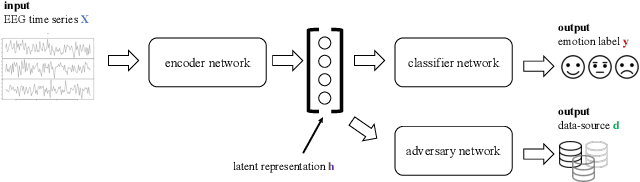
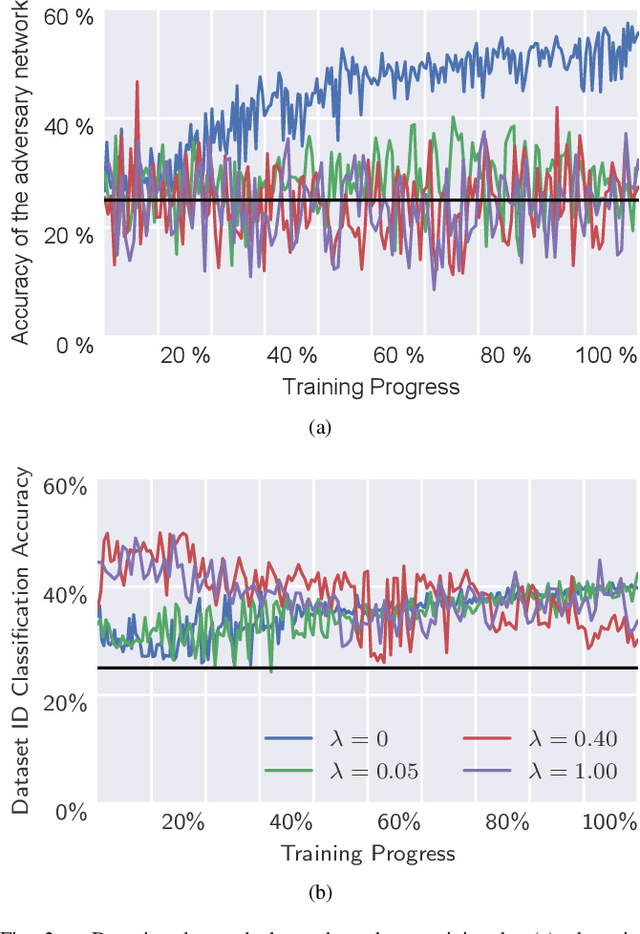
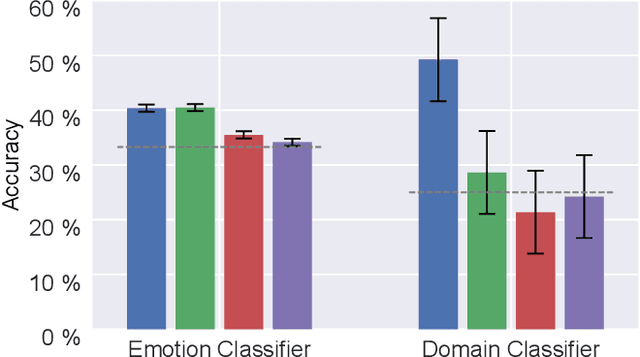
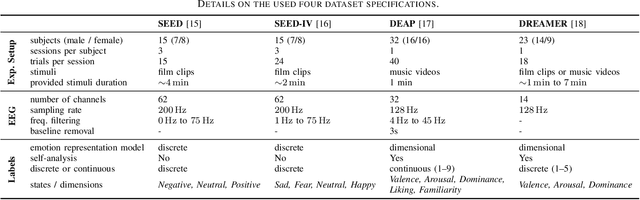
Abstract:Electroencephalography (EEG) is shown to be a valuable data source for evaluating subjects' mental states. However, the interpretation of multi-modal EEG signals is challenging, as they suffer from poor signal-to-noise-ratio, are highly subject-dependent, and are bound to the equipment and experimental setup used, (i.e. domain). This leads to machine learning models often suffer from poor generalization ability, where they perform significantly worse on real-world data than on the exploited training data. Recent research heavily focuses on cross-subject and cross-session transfer learning frameworks to reduce domain calibration efforts for EEG signals. We argue that multi-source learning via learning domain-invariant representations from multiple data-sources is a viable alternative, as the available data from different EEG data-source domains (e.g., subjects, sessions, experimental setups) grow massively. We propose an adversarial inference approach to learn data-source invariant representations in this context, enabling multi-source learning for EEG-based brain-computer interfaces. We unify EEG recordings from different source domains (i.e., emotion recognition datasets SEED, SEED-IV, DEAP, DREAMER), and demonstrate the feasibility of our invariant representation learning approach in suppressing data-source-relevant information leakage by 35% while still achieving stable EEG-based emotion classification performance.
Domain-Invariant Representation Learning from EEG with Private Encoders
Jan 27, 2022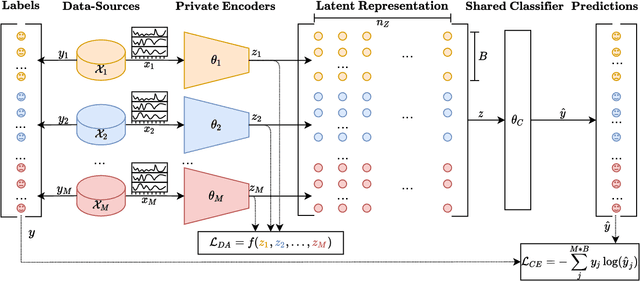
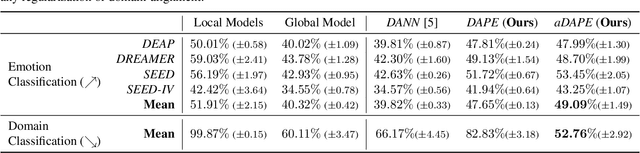
Abstract:Deep learning based electroencephalography (EEG) signal processing methods are known to suffer from poor test-time generalization due to the changes in data distribution. This becomes a more challenging problem when privacy-preserving representation learning is of interest such as in clinical settings. To that end, we propose a multi-source learning architecture where we extract domain-invariant representations from dataset-specific private encoders. Our model utilizes a maximum-mean-discrepancy (MMD) based domain alignment approach to impose domain-invariance for encoded representations, which outperforms state-of-the-art approaches in EEG-based emotion classification. Furthermore, representations learned in our pipeline preserve domain privacy as dataset-specific private encoding alleviates the need for conventional, centralized EEG-based deep neural network training approaches with shared parameters.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge