Nikolaos Nakis
Machine Learning for Static and Single-Event Dynamic Complex Network Analysis
Dec 19, 2025Abstract:The primary objective of this thesis is to develop novel algorithmic approaches for Graph Representation Learning of static and single-event dynamic networks. In such a direction, we focus on the family of Latent Space Models, and more specifically on the Latent Distance Model which naturally conveys important network characteristics such as homophily, transitivity, and the balance theory. Furthermore, this thesis aims to create structural-aware network representations, which lead to hierarchical expressions of network structure, community characterization, the identification of extreme profiles in networks, and impact dynamics quantification in temporal networks. Crucially, the methods presented are designed to define unified learning processes, eliminating the need for heuristics and multi-stage processes like post-processing steps. Our aim is to delve into a journey towards unified network embeddings that are both comprehensive and powerful, capable of characterizing network structures and adeptly handling the diverse tasks that graph analysis offers.
The Signed Two-Space Proximity Model for Learning Representations in Protein-Protein Interaction Networks
Mar 05, 2025Abstract:Accurately predicting complex protein-protein interactions (PPIs) is crucial for decoding biological processes, from cellular functioning to disease mechanisms. However, experimental methods for determining PPIs are computationally expensive. Thus, attention has been recently drawn to machine learning approaches. Furthermore, insufficient effort has been made toward analyzing signed PPI networks, which capture both activating (positive) and inhibitory (negative) interactions. To accurately represent biological relationships, we present the Signed Two-Space Proximity Model (S2-SPM) for signed PPI networks, which explicitly incorporates both types of interactions, reflecting the complex regulatory mechanisms within biological systems. This is achieved by leveraging two independent latent spaces to differentiate between positive and negative interactions while representing protein similarity through proximity in these spaces. Our approach also enables the identification of archetypes representing extreme protein profiles. S2-SPM's superior performance in predicting the presence and sign of interactions in SPPI networks is demonstrated in link prediction tasks against relevant baseline methods. Additionally, the biological prevalence of the identified archetypes is confirmed by an enrichment analysis of Gene Ontology (GO) terms, which reveals that distinct biological tasks are associated with archetypal groups formed by both interactions. This study is also validated regarding statistical significance and sensitivity analysis, providing insights into the functional roles of different interaction types. Finally, the robustness and consistency of the extracted archetype structures are confirmed using the Bayesian Normalized Mutual Information (BNMI) metric, proving the model's reliability in capturing meaningful SPPI patterns.
How Low Can You Go? Searching for the Intrinsic Dimensionality of Complex Networks using Metric Node Embeddings
Mar 03, 2025



Abstract:Low-dimensional embeddings are essential for machine learning tasks involving graphs, such as node classification, link prediction, community detection, network visualization, and network compression. Although recent studies have identified exact low-dimensional embeddings, the limits of the required embedding dimensions remain unclear. We presently prove that lower dimensional embeddings are possible when using Euclidean metric embeddings as opposed to vector-based Logistic PCA (LPCA) embeddings. In particular, we provide an efficient logarithmic search procedure for identifying the exact embedding dimension and demonstrate how metric embeddings enable inference of the exact embedding dimensions of large-scale networks by exploiting that the metric properties can be used to provide linearithmic scaling. Empirically, we show that our approach extracts substantially lower dimensional representations of networks than previously reported for small-sized networks. For the first time, we demonstrate that even large-scale networks can be effectively embedded in very low-dimensional spaces, and provide examples of scalable, exact reconstruction for graphs with up to a million nodes. Our approach highlights that the intrinsic dimensionality of networks is substantially lower than previously reported and provides a computationally efficient assessment of the exact embedding dimension also of large-scale networks. The surprisingly low dimensional representations achieved demonstrate that networks in general can be losslessly represented using very low dimensional feature spaces, which can be used to guide existing network analysis tasks from community detection and node classification to structure revealing exact network visualizations.
Signed Graph Autoencoder for Explainable and Polarization-Aware Network Embeddings
Sep 16, 2024

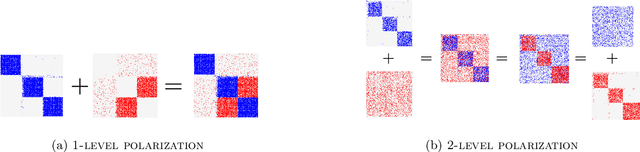

Abstract:Autoencoders based on Graph Neural Networks (GNNs) have garnered significant attention in recent years for their ability to extract informative latent representations, characterizing the structure of complex topologies, such as graphs. Despite the prevalence of Graph Autoencoders, there has been limited focus on developing and evaluating explainable neural-based graph generative models specifically designed for signed networks. To address this gap, we propose the Signed Graph Archetypal Autoencoder (SGAAE) framework. SGAAE extracts node-level representations that express node memberships over distinct extreme profiles, referred to as archetypes, within the network. This is achieved by projecting the graph onto a learned polytope, which governs its polarization. The framework employs a recently proposed likelihood for analyzing signed networks based on the Skellam distribution, combined with relational archetypal analysis and GNNs. Our experimental evaluation demonstrates the SGAAEs' capability to successfully infer node memberships over the different underlying latent structures while extracting competing communities formed through the participation of the opposing views in the network. Additionally, we introduce the 2-level network polarization problem and show how SGAAE is able to characterize such a setting. The proposed model achieves high performance in different tasks of signed link prediction across four real-world datasets, outperforming several baseline models.
Time to Cite: Modeling Citation Networks using the Dynamic Impact Single-Event Embedding Model
Feb 28, 2024Abstract:Understanding the structure and dynamics of scientific research, i.e., the science of science (SciSci), has become an important area of research in order to address imminent questions including how scholars interact to advance science, how disciplines are related and evolve, and how research impact can be quantified and predicted. Central to the study of SciSci has been the analysis of citation networks. Here, two prominent modeling methodologies have been employed: one is to assess the citation impact dynamics of papers using parametric distributions, and the other is to embed the citation networks in a latent space optimal for characterizing the static relations between papers in terms of their citations. Interestingly, citation networks are a prominent example of single-event dynamic networks, i.e., networks for which each dyad only has a single event (i.e., the point in time of citation). We presently propose a novel likelihood function for the characterization of such single-event networks. Using this likelihood, we propose the Dynamic Impact Single-Event Embedding model (DISEE). The \textsc{\modelabbrev} model characterizes the scientific interactions in terms of a latent distance model in which random effects account for citation heterogeneity while the time-varying impact is characterized using existing parametric representations for assessment of dynamic impact. We highlight the proposed approach on several real citation networks finding that the DISEE well reconciles static latent distance network embedding approaches with classical dynamic impact assessments.
Continuous-time Graph Representation with Sequential Survival Process
Dec 20, 2023Abstract:Over the past two decades, there has been a tremendous increase in the growth of representation learning methods for graphs, with numerous applications across various fields, including bioinformatics, chemistry, and the social sciences. However, current dynamic network approaches focus on discrete-time networks or treat links in continuous-time networks as instantaneous events. Therefore, these approaches have limitations in capturing the persistence or absence of links that continuously emerge and disappear over time for particular durations. To address this, we propose a novel stochastic process relying on survival functions to model the durations of links and their absences over time. This forms a generic new likelihood specification explicitly accounting for intermittent edge-persistent networks, namely GraSSP: Graph Representation with Sequential Survival Process. We apply the developed framework to a recent continuous time dynamic latent distance model characterizing network dynamics in terms of a sequence of piecewise linear movements of nodes in latent space. We quantitatively assess the developed framework in various downstream tasks, such as link prediction and network completion, demonstrating that the developed modeling framework accounting for link persistence and absence well tracks the intrinsic trajectories of nodes in a latent space and captures the underlying characteristics of evolving network structure.
A Hybrid Membership Latent Distance Model for Unsigned and Signed Integer Weighted Networks
Aug 29, 2023Abstract:Graph representation learning (GRL) has become a prominent tool for furthering the understanding of complex networks providing tools for network embedding, link prediction, and node classification. In this paper, we propose the Hybrid Membership-Latent Distance Model (HM-LDM) by exploring how a Latent Distance Model (LDM) can be constrained to a latent simplex. By controlling the edge lengths of the corners of the simplex, the volume of the latent space can be systematically controlled. Thereby communities are revealed as the space becomes more constrained, with hard memberships being recovered as the simplex volume goes to zero. We further explore a recent likelihood formulation for signed networks utilizing the Skellam distribution to account for signed weighted networks and extend the HM-LDM to the signed Hybrid Membership-Latent Distance Model (sHM-LDM). Importantly, the induced likelihood function explicitly attracts nodes with positive links and deters nodes from having negative interactions. We demonstrate the utility of HM-LDM and sHM-LDM on several real networks. We find that the procedures successfully identify prominent distinct structures, as well as how nodes relate to the extracted aspects providing favorable performances in terms of link prediction when compared to prominent baselines. Furthermore, the learned soft memberships enable easily interpretable network visualizations highlighting distinct patterns.
Characterizing Polarization in Social Networks using the Signed Relational Latent Distance Model
Jan 28, 2023Abstract:Graph representation learning has become a prominent tool for the characterization and understanding of the structure of networks in general and social networks in particular. Typically, these representation learning approaches embed the networks into a low-dimensional space in which the role of each individual can be characterized in terms of their latent position. A major current concern in social networks is the emergence of polarization and filter bubbles promoting a mindset of "us-versus-them" that may be defined by extreme positions believed to ultimately lead to political violence and the erosion of democracy. Such polarized networks are typically characterized in terms of signed links reflecting likes and dislikes. We propose the latent Signed relational Latent dIstance Model (SLIM) utilizing for the first time the Skellam distribution as a likelihood function for signed networks and extend the modeling to the characterization of distinct extreme positions by constraining the embedding space to polytopes. On four real social signed networks of polarization, we demonstrate that the model extracts low-dimensional characterizations that well predict friendships and animosity while providing interpretable visualizations defined by extreme positions when endowing the model with an embedding space restricted to polytopes.
Piecewise-Velocity Model for Learning Continuous-time Dynamic Node Representations
Dec 23, 2022



Abstract:Networks have become indispensable and ubiquitous structures in many fields to model the interactions among different entities, such as friendship in social networks or protein interactions in biological graphs. A major challenge is to understand the structure and dynamics of these systems. Although networks evolve through time, most existing graph representation learning methods target only static networks. Whereas approaches have been developed for the modeling of dynamic networks, there is a lack of efficient continuous time dynamic graph representation learning methods that can provide accurate network characterization and visualization in low dimensions while explicitly accounting for prominent network characteristics such as homophily and transitivity. In this paper, we propose the Piecewise-Velocity Model (PiVeM) for the representation of continuous-time dynamic networks. It learns dynamic embeddings in which the temporal evolution of nodes is approximated by piecewise linear interpolations based on a latent distance model with piecewise constant node-specific velocities. The model allows for analytically tractable expressions of the associated Poisson process likelihood with scalable inference invariant to the number of events. We further impose a scalable Kronecker structured Gaussian Process prior to the dynamics accounting for community structure, temporal smoothness, and disentangled (uncorrelated) latent embedding dimensions optimally learned to characterize the network dynamics. We show that PiVeM can successfully represent network structure and dynamics in ultra-low two-dimensional spaces. It outperforms relevant state-of-art methods in downstream tasks such as link prediction. In summary, PiVeM enables easily interpretable dynamic network visualizations and characterizations that can further improve our understanding of the intrinsic dynamics of time-evolving networks.
A Hierarchical Block Distance Model for Ultra Low-Dimensional Graph Representations
Apr 12, 2022



Abstract:Graph Representation Learning (GRL) has become central for characterizing structures of complex networks and performing tasks such as link prediction, node classification, network reconstruction, and community detection. Whereas numerous generative GRL models have been proposed, many approaches have prohibitive computational requirements hampering large-scale network analysis, fewer are able to explicitly account for structure emerging at multiple scales, and only a few explicitly respect important network properties such as homophily and transitivity. This paper proposes a novel scalable graph representation learning method named the Hierarchical Block Distance Model (HBDM). The HBDM imposes a multiscale block structure akin to stochastic block modeling (SBM) and accounts for homophily and transitivity by accurately approximating the latent distance model (LDM) throughout the inferred hierarchy. The HBDM naturally accommodates unipartite, directed, and bipartite networks whereas the hierarchy is designed to ensure linearithmic time and space complexity enabling the analysis of very large-scale networks. We evaluate the performance of the HBDM on massive networks consisting of millions of nodes. Importantly, we find that the proposed HBDM framework significantly outperforms recent scalable approaches in all considered downstream tasks. Surprisingly, we observe superior performance even imposing ultra-low two-dimensional embeddings facilitating accurate direct and hierarchical-aware network visualization and interpretation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge