Abdulkadir Celikkanat
Revisiting K-mer Profile for Effective and Scalable Genome Representation Learning
Nov 04, 2024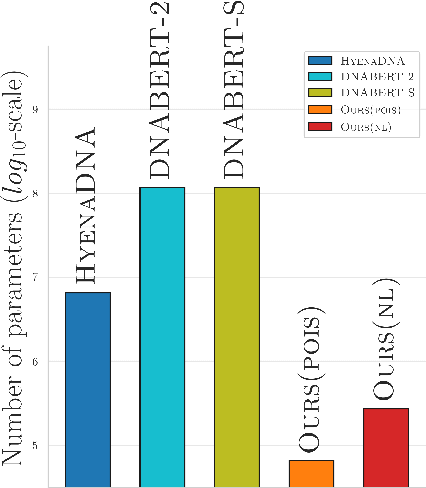

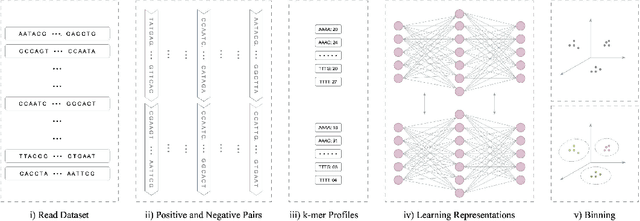
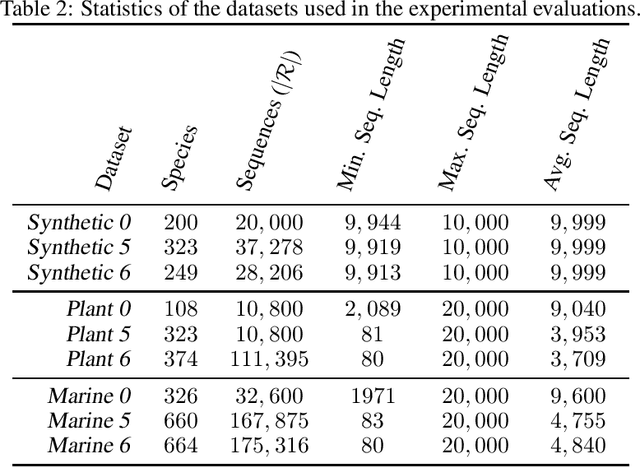
Abstract:Obtaining effective representations of DNA sequences is crucial for genome analysis. Metagenomic binning, for instance, relies on genome representations to cluster complex mixtures of DNA fragments from biological samples with the aim of determining their microbial compositions. In this paper, we revisit k-mer-based representations of genomes and provide a theoretical analysis of their use in representation learning. Based on the analysis, we propose a lightweight and scalable model for performing metagenomic binning at the genome read level, relying only on the k-mer compositions of the DNA fragments. We compare the model to recent genome foundation models and demonstrate that while the models are comparable in performance, the proposed model is significantly more effective in terms of scalability, a crucial aspect for performing metagenomic binning of real-world datasets.
Time to Cite: Modeling Citation Networks using the Dynamic Impact Single-Event Embedding Model
Feb 28, 2024Abstract:Understanding the structure and dynamics of scientific research, i.e., the science of science (SciSci), has become an important area of research in order to address imminent questions including how scholars interact to advance science, how disciplines are related and evolve, and how research impact can be quantified and predicted. Central to the study of SciSci has been the analysis of citation networks. Here, two prominent modeling methodologies have been employed: one is to assess the citation impact dynamics of papers using parametric distributions, and the other is to embed the citation networks in a latent space optimal for characterizing the static relations between papers in terms of their citations. Interestingly, citation networks are a prominent example of single-event dynamic networks, i.e., networks for which each dyad only has a single event (i.e., the point in time of citation). We presently propose a novel likelihood function for the characterization of such single-event networks. Using this likelihood, we propose the Dynamic Impact Single-Event Embedding model (DISEE). The \textsc{\modelabbrev} model characterizes the scientific interactions in terms of a latent distance model in which random effects account for citation heterogeneity while the time-varying impact is characterized using existing parametric representations for assessment of dynamic impact. We highlight the proposed approach on several real citation networks finding that the DISEE well reconciles static latent distance network embedding approaches with classical dynamic impact assessments.
Continuous-time Graph Representation with Sequential Survival Process
Dec 20, 2023Abstract:Over the past two decades, there has been a tremendous increase in the growth of representation learning methods for graphs, with numerous applications across various fields, including bioinformatics, chemistry, and the social sciences. However, current dynamic network approaches focus on discrete-time networks or treat links in continuous-time networks as instantaneous events. Therefore, these approaches have limitations in capturing the persistence or absence of links that continuously emerge and disappear over time for particular durations. To address this, we propose a novel stochastic process relying on survival functions to model the durations of links and their absences over time. This forms a generic new likelihood specification explicitly accounting for intermittent edge-persistent networks, namely GraSSP: Graph Representation with Sequential Survival Process. We apply the developed framework to a recent continuous time dynamic latent distance model characterizing network dynamics in terms of a sequence of piecewise linear movements of nodes in latent space. We quantitatively assess the developed framework in various downstream tasks, such as link prediction and network completion, demonstrating that the developed modeling framework accounting for link persistence and absence well tracks the intrinsic trajectories of nodes in a latent space and captures the underlying characteristics of evolving network structure.
Multiple Kernel Representation Learning on Networks
Jun 09, 2021



Abstract:Learning representations of nodes in a low dimensional space is a crucial task with numerous interesting applications in network analysis, including link prediction, node classification, and visualization. Two popular approaches for this problem are matrix factorization and random walk-based models. In this paper, we aim to bring together the best of both worlds, towards learning node representations. In particular, we propose a weighted matrix factorization model that encodes random walk-based information about nodes of the network. The benefit of this novel formulation is that it enables us to utilize kernel functions without realizing the exact proximity matrix so that it enhances the expressiveness of existing matrix decomposition methods with kernels and alleviates their computational complexities. We extend the approach with a multiple kernel learning formulation that provides the flexibility of learning the kernel as the linear combination of a dictionary of kernels in data-driven fashion. We perform an empirical evaluation on real-world networks, showing that the proposed model outperforms baseline node embedding algorithms in downstream machine learning tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge