Nati Daniel
CSG: A Context-Semantic Guided Diffusion Approach in De Novo Musculoskeletal Ultrasound Image Generation
Dec 08, 2024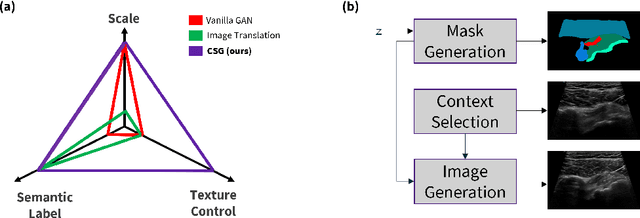
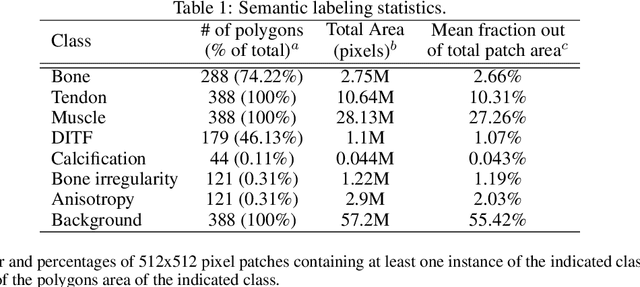
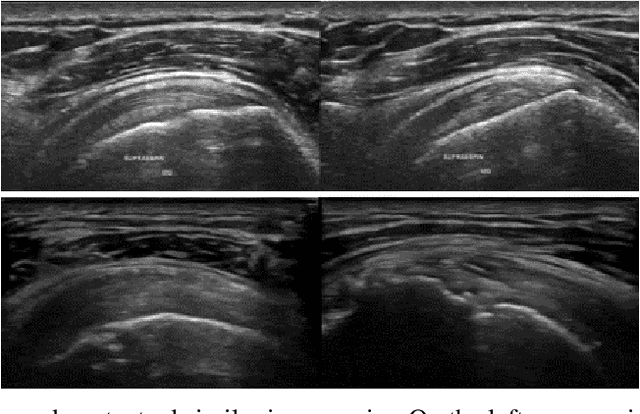
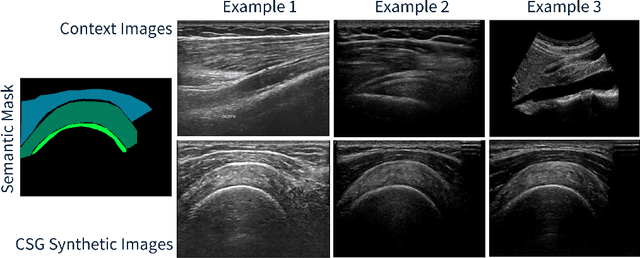
Abstract:The use of synthetic images in medical imaging Artificial Intelligence (AI) solutions has been shown to be beneficial in addressing the limited availability of diverse, unbiased, and representative data. Despite the extensive use of synthetic image generation methods, controlling the semantics variability and context details remains challenging, limiting their effectiveness in producing diverse and representative medical image datasets. In this work, we introduce a scalable semantic and context-conditioned generative model, coined CSG (Context-Semantic Guidance). This dual conditioning approach allows for comprehensive control over both structure and appearance, advancing the synthesis of realistic and diverse ultrasound images. We demonstrate the ability of CSG to generate findings (pathological anomalies) in musculoskeletal (MSK) ultrasound images. Moreover, we test the quality of the synthetic images using a three-fold validation protocol. The results show that the synthetic images generated by CSG improve the performance of semantic segmentation models, exhibit enhanced similarity to real images compared to the baseline methods, and are undistinguishable from real images according to a Turing test. Furthermore, we demonstrate an extension of the CSG that allows enhancing the variability space of images by synthetically generating augmentations of anatomical geometries and textures.
PriorPath: Coarse-To-Fine Approach for Controlled De-Novo Pathology Semantic Masks Generation
Nov 25, 2024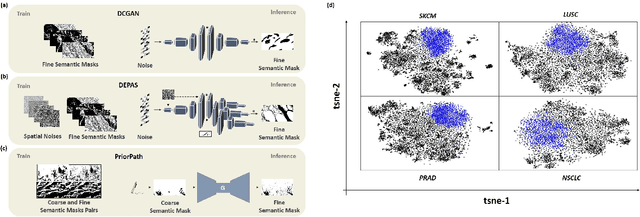
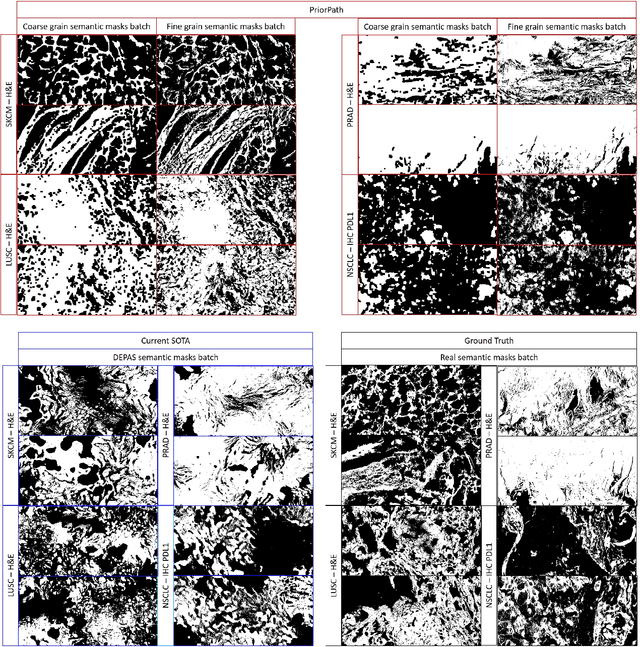
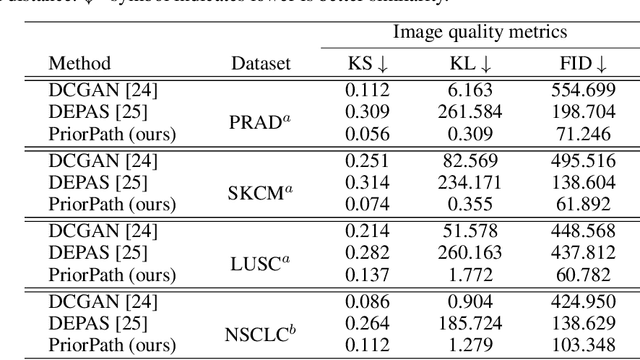
Abstract:Incorporating artificial intelligence (AI) into digital pathology offers promising prospects for automating and enhancing tasks such as image analysis and diagnostic processes. However, the diversity of tissue samples and the necessity for meticulous image labeling often result in biased datasets, constraining the applicability of algorithms trained on them. To harness synthetic histopathological images to cope with this challenge, it is essential not only to produce photorealistic images but also to be able to exert control over the cellular characteristics they depict. Previous studies used methods to generate, from random noise, semantic masks that captured the spatial distribution of the tissue. These masks were then used as a prior for conditional generative approaches to produce photorealistic histopathological images. However, as with many other generative models, this solution exhibits mode collapse as the model fails to capture the full diversity of the underlying data distribution. In this work, we present a pipeline, coined PriorPath, that generates detailed, realistic, semantic masks derived from coarse-grained images delineating tissue regions. This approach enables control over the spatial arrangement of the generated masks and, consequently, the resulting synthetic images. We demonstrated the efficacy of our method across three cancer types, skin, prostate, and lung, showcasing PriorPath's capability to cover the semantic mask space and to provide better similarity to real masks compared to previous methods. Our approach allows for specifying desired tissue distributions and obtaining both photorealistic masks and images within a single platform, thus providing a state-of-the-art, controllable solution for generating histopathological images to facilitate AI for computational pathology.
Semantic Segmentation Refiner for Ultrasound Applications with Zero-Shot Foundation Models
Apr 25, 2024Abstract:Despite the remarkable success of deep learning in medical imaging analysis, medical image segmentation remains challenging due to the scarcity of high-quality labeled images for supervision. Further, the significant domain gap between natural and medical images in general and ultrasound images in particular hinders fine-tuning models trained on natural images to the task at hand. In this work, we address the performance degradation of segmentation models in low-data regimes and propose a prompt-less segmentation method harnessing the ability of segmentation foundation models to segment abstract shapes. We do that via our novel prompt point generation algorithm which uses coarse semantic segmentation masks as input and a zero-shot prompt-able foundation model as an optimization target. We demonstrate our method on a segmentation findings task (pathologic anomalies) in ultrasound images. Our method's advantages are brought to light in varying degrees of low-data regime experiments on a small-scale musculoskeletal ultrasound images dataset, yielding a larger performance gain as the training set size decreases.
Facial Expression Re-targeting from a Single Character
Jun 21, 2023



Abstract:Video retargeting for digital face animation is used in virtual reality, social media, gaming, movies, and video conference, aiming to animate avatars' facial expressions based on videos of human faces. The standard method to represent facial expressions for 3D characters is by blendshapes, a vector of weights representing the avatar's neutral shape and its variations under facial expressions, e.g., smile, puff, blinking. Datasets of paired frames with blendshape vectors are rare, and labeling can be laborious, time-consuming, and subjective. In this work, we developed an approach that handles the lack of appropriate datasets. Instead, we used a synthetic dataset of only one character. To generalize various characters, we re-represented each frame to face landmarks. We developed a unique deep-learning architecture that groups landmarks for each facial organ and connects them to relevant blendshape weights. Additionally, we incorporated complementary methods for facial expressions that landmarks did not represent well and gave special attention to eye expressions. We have demonstrated the superiority of our approach to previous research in qualitative and quantitative metrics. Our approach achieved a higher MOS of 68% and a lower MSE of 44.2% when tested on videos with various users and expressions.
Between Generating Noise and Generating Images: Noise in the Correct Frequency Improves the Quality of Synthetic Histopathology Images for Digital Pathology
Feb 13, 2023



Abstract:Artificial intelligence and machine learning techniques have the promise to revolutionize the field of digital pathology. However, these models demand considerable amounts of data, while the availability of unbiased training data is limited. Synthetic images can augment existing datasets, to improve and validate AI algorithms. Yet, controlling the exact distribution of cellular features within them is still challenging. One of the solutions is harnessing conditional generative adversarial networks that take a semantic mask as an input rather than a random noise. Unlike other domains, outlining the exact cellular structure of tissues is hard, and most of the input masks depict regions of cell types. However, using polygon-based masks introduce inherent artifacts within the synthetic images - due to the mismatch between the polygon size and the single-cell size. In this work, we show that introducing random single-pixel noise with the appropriate spatial frequency into a polygon semantic mask can dramatically improve the quality of the synthetic images. We used our platform to generate synthetic images of immunohistochemistry-treated lung biopsies. We test the quality of the images using a three-fold validation procedure. First, we show that adding the appropriate noise frequency yields 87% of the similarity metrics improvement that is obtained by adding the actual single-cell features. Second, we show that the synthetic images pass the Turing test. Finally, we show that adding these synthetic images to the train set improves AI performance in terms of PD-L1 semantic segmentation performances. Our work suggests a simple and powerful approach for generating synthetic data on demand to unbias limited datasets to improve the algorithms' accuracy and validate their robustness.
DEPAS: De-novo Pathology Semantic Masks using a Generative Model
Feb 13, 2023
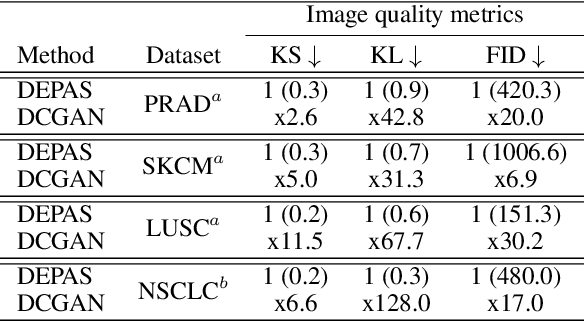
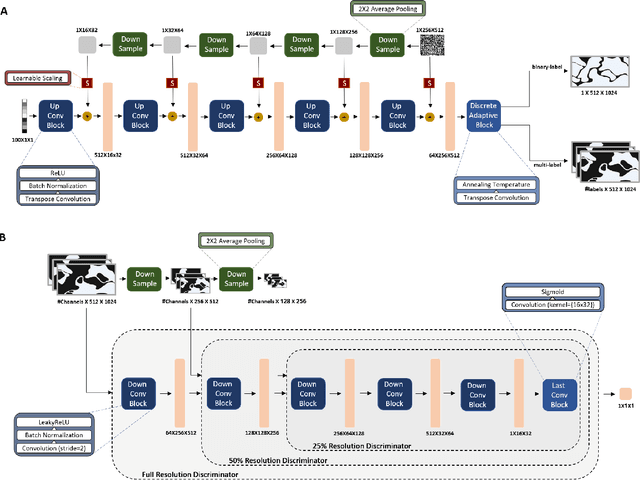
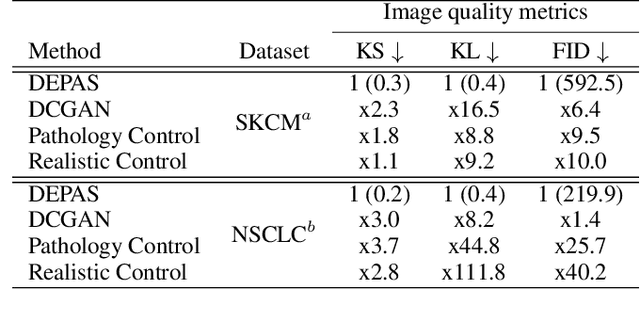
Abstract:The integration of artificial intelligence into digital pathology has the potential to automate and improve various tasks, such as image analysis and diagnostic decision-making. Yet, the inherent variability of tissues, together with the need for image labeling, lead to biased datasets that limit the generalizability of algorithms trained on them. One of the emerging solutions for this challenge is synthetic histological images. However, debiasing real datasets require not only generating photorealistic images but also the ability to control the features within them. A common approach is to use generative methods that perform image translation between semantic masks that reflect prior knowledge of the tissue and a histological image. However, unlike other image domains, the complex structure of the tissue prevents a simple creation of histology semantic masks that are required as input to the image translation model, while semantic masks extracted from real images reduce the process's scalability. In this work, we introduce a scalable generative model, coined as DEPAS, that captures tissue structure and generates high-resolution semantic masks with state-of-the-art quality. We demonstrate the ability of DEPAS to generate realistic semantic maps of tissue for three types of organs: skin, prostate, and lung. Moreover, we show that these masks can be processed using a generative image translation model to produce photorealistic histology images of two types of cancer with two different types of staining techniques. Finally, we harness DEPAS to generate multi-label semantic masks that capture different cell types distributions and use them to produce histological images with on-demand cellular features. Overall, our work provides a state-of-the-art solution for the challenging task of generating synthetic histological images while controlling their semantic information in a scalable way.
Harnessing Artificial Intelligence to Infer Novel Spatial Biomarkers for the Diagnosis of Eosinophilic Esophagitis
May 26, 2022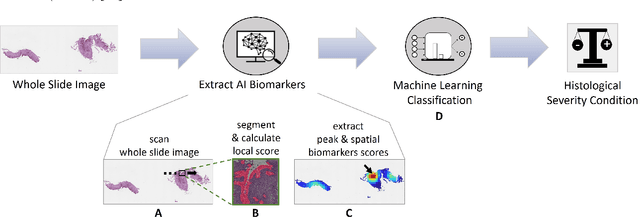
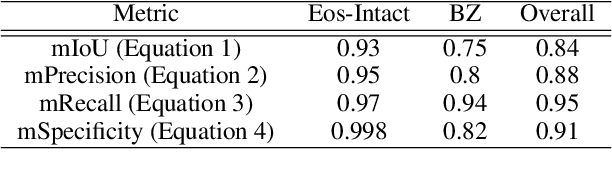
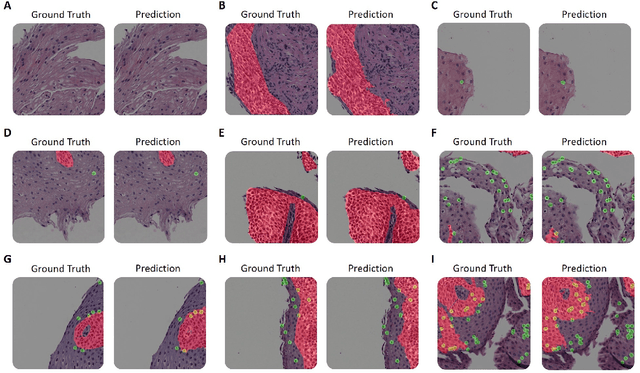
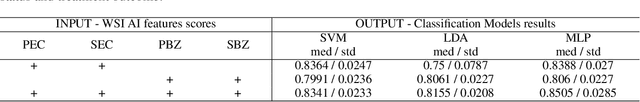
Abstract:Eosinophilic esophagitis (EoE) is a chronic allergic inflammatory condition of the esophagus associated with elevated esophageal eosinophils. Second only to gastroesophageal reflux disease, EoE is one of the leading causes of chronic refractory dysphagia in adults and children. EoE diagnosis requires enumerating the density of esophageal eosinophils in esophageal biopsies, a somewhat subjective task that is time-consuming, thus reducing the ability to process the complex tissue structure. Previous artificial intelligence (AI) approaches that aimed to improve histology-based diagnosis focused on recapitulating identification and quantification of the area of maximal eosinophil density. However, this metric does not account for the distribution of eosinophils or other histological features, over the whole slide image. Here, we developed an artificial intelligence platform that infers local and spatial biomarkers based on semantic segmentation of intact eosinophils and basal zone distributions. Besides the maximal density of eosinophils (referred to as Peak Eosinophil Count [PEC]) and a maximal basal zone fraction, we identify two additional metrics that reflect the distribution of eosinophils and basal zone fractions. This approach enables a decision support system that predicts EoE activity and classifies the histological severity of EoE patients. We utilized a cohort that includes 1066 biopsy slides from 400 subjects to validate the system's performance and achieved a histological severity classification accuracy of 86.70%, sensitivity of 84.50%, and specificity of 90.09%. Our approach highlights the importance of systematically analyzing the distribution of biopsy features over the entire slide and paves the way towards a personalized decision support system that will assist not only in counting cells but can also potentially improve diagnosis and provide treatment prediction.
Crowd Source Scene Change Detection and Local Map Update
Mar 10, 2022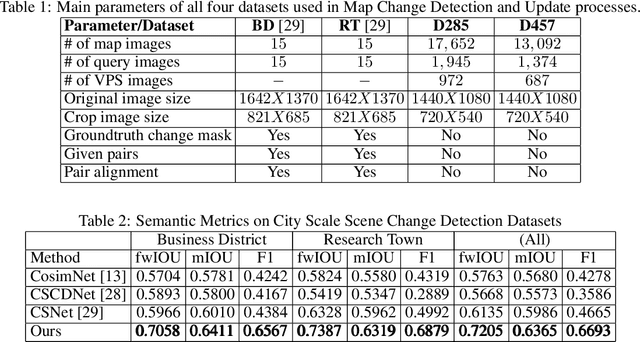


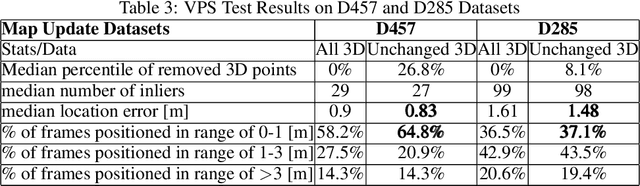
Abstract:As scene changes with time map descriptors become outdated, affecting VPS localization accuracy. In this work, we propose an approach to detect structural and texture scene changes to be followed by map update. In our method - map includes 3D points with descriptors generated either via LiDAR or SFM. Common approaches suffer from shortcomings: 1) Direct comparison of the two point-clouds for change detection is slow due to the need to build new point-cloud every time we want to compare; 2) Image based comparison requires to keep the map images adding substantial storage overhead. To circumvent this problems, we propose an approach based on point-clouds descriptors comparison: 1) Based on VPS poses select close query and map images pairs, 2) Registration of query images to map image descriptors, 3) Use segmentation to filter out dynamic or short term temporal changes, 4) Compare the descriptors between corresponding segments.
PECNet: A Deep Multi-Label Segmentation Network for Eosinophilic Esophagitis Biopsy Diagnostics
Mar 02, 2021
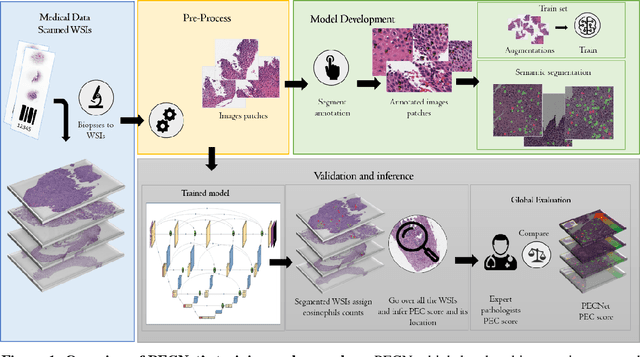
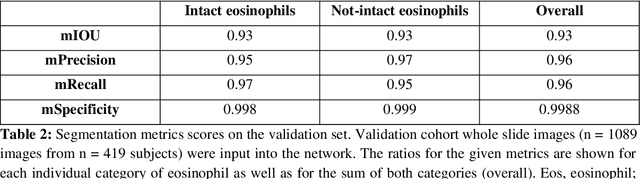

Abstract:Background. Eosinophilic esophagitis (EoE) is an allergic inflammatory condition of the esophagus associated with elevated numbers of eosinophils. Disease diagnosis and monitoring requires determining the concentration of eosinophils in esophageal biopsies, a time-consuming, tedious and somewhat subjective task currently performed by pathologists. Methods. Herein, we aimed to use machine learning to identify, quantitate and diagnose EoE. We labeled more than 100M pixels of 4345 images obtained by scanning whole slides of H&E-stained sections of esophageal biopsies derived from 23 EoE patients. We used this dataset to train a multi-label segmentation deep network. To validate the network, we examined a replication cohort of 1089 whole slide images from 419 patients derived from multiple institutions. Findings. PECNet segmented both intact and not-intact eosinophils with a mean intersection over union (mIoU) of 0.93. This segmentation was able to quantitate intact eosinophils with a mean absolute error of 0.611 eosinophils and classify EoE disease activity with an accuracy of 98.5%. Using whole slide images from the validation cohort, PECNet achieved an accuracy of 94.8%, sensitivity of 94.3%, and specificity of 95.14% in reporting EoE disease activity. Interpretation. We have developed a deep learning multi-label semantic segmentation network that successfully addresses two of the main challenges in EoE diagnostics and digital pathology, the need to detect several types of small features simultaneously and the ability to analyze whole slides efficiently. Our results pave the way for an automated diagnosis of EoE and can be utilized for other conditions with similar challenges.
Machine learning approach for biopsy-based identification of eosinophilic esophagitis reveals importance of global features
Jan 13, 2021Abstract:Goal: Eosinophilic esophagitis (EoE) is an allergic inflammatory condition characterized by eosinophil accumulation in the esophageal mucosa. EoE diagnosis includes a manual assessment of eosinophil levels in mucosal biopsies - a time-consuming, laborious task that is difficult to standardize. One of the main challenges in automating this process, like many other biopsy-based diagnostics, is detecting features that are small relative to the size of the biopsy. Results: In this work, we utilized hematoxylin- and eosin-stained slides from esophageal biopsies from patients with active EoE and control subjects to develop a platform based on a deep convolutional neural network (DCNN) that can classify esophageal biopsies with an accuracy of 85%, sensitivity of 82.5%, and specificity of 87%. Moreover, by combining several downscaling and cropping strategies, we show that some of the features contributing to the correct classification are global rather than specific, local features. Conclusions: We report the ability of artificial intelligence to identify EoE using computer vision analysis of esophageal biopsy slides. Further, the DCNN features associated with EoE are based on not only local eosinophils but also global histologic changes. Our approach can be used for other conditions that rely on biopsy-based histologic diagnostics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge