Marc E. Rothenberg
Harnessing Digital Pathology And Causal Learning To Improve Eosinophilic Esophagitis Dietary Treatment Assignment
Apr 16, 2023



Abstract:Eosinophilic esophagitis (EoE) is a chronic, food antigen-driven, allergic inflammatory condition of the esophagus associated with elevated esophageal eosinophils. EoE is a top cause of chronic dysphagia after GERD. Diagnosis of EoE relies on counting eosinophils in histological slides, a manual and time-consuming task that limits the ability to extract complex patient-dependent features. The treatment of EoE includes medication and food elimination. A personalized food elimination plan is crucial for engagement and efficiency, but previous attempts failed to produce significant results. In this work, on the one hand, we utilize AI for inferring histological features from the entire biopsy slide, features that cannot be extracted manually. On the other hand, we develop causal learning models that can process this wealth of data. We applied our approach to the 'Six-Food vs. One-Food Eosinophilic Esophagitis Diet Study', where 112 symptomatic adults aged 18-60 years with active EoE were assigned to either a six-food elimination diet (6FED) or a one-food elimination diet (1FED) for six weeks. Our results show that the average treatment effect (ATE) of the 6FED treatment compared with the 1FED treatment is not significant, that is, neither diet was superior to the other. We examined several causal models and show that the best treatment strategy was obtained using T-learner with two XGBoost modules. While 1FED only and 6FED only provide improvement for 35%-38% of the patients, which is not significantly different from a random treatment assignment, our causal model yields a significantly better improvement rate of 58.4%. This study illustrates the significance of AI in enhancing treatment planning by analyzing molecular features' distribution in histological slides through causal learning. Our approach can be harnessed for other conditions that rely on histology for diagnosis and treatment.
Harnessing Artificial Intelligence to Infer Novel Spatial Biomarkers for the Diagnosis of Eosinophilic Esophagitis
May 26, 2022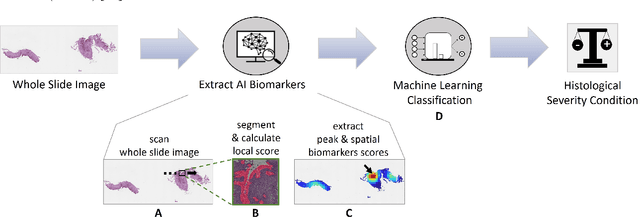
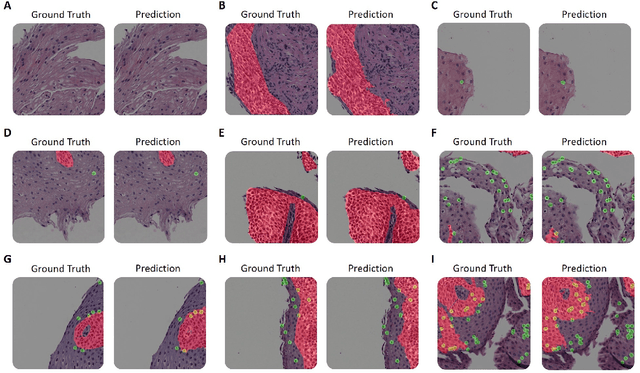
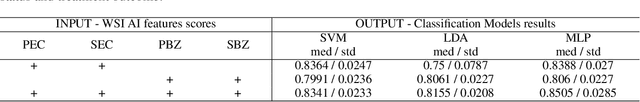
Abstract:Eosinophilic esophagitis (EoE) is a chronic allergic inflammatory condition of the esophagus associated with elevated esophageal eosinophils. Second only to gastroesophageal reflux disease, EoE is one of the leading causes of chronic refractory dysphagia in adults and children. EoE diagnosis requires enumerating the density of esophageal eosinophils in esophageal biopsies, a somewhat subjective task that is time-consuming, thus reducing the ability to process the complex tissue structure. Previous artificial intelligence (AI) approaches that aimed to improve histology-based diagnosis focused on recapitulating identification and quantification of the area of maximal eosinophil density. However, this metric does not account for the distribution of eosinophils or other histological features, over the whole slide image. Here, we developed an artificial intelligence platform that infers local and spatial biomarkers based on semantic segmentation of intact eosinophils and basal zone distributions. Besides the maximal density of eosinophils (referred to as Peak Eosinophil Count [PEC]) and a maximal basal zone fraction, we identify two additional metrics that reflect the distribution of eosinophils and basal zone fractions. This approach enables a decision support system that predicts EoE activity and classifies the histological severity of EoE patients. We utilized a cohort that includes 1066 biopsy slides from 400 subjects to validate the system's performance and achieved a histological severity classification accuracy of 86.70%, sensitivity of 84.50%, and specificity of 90.09%. Our approach highlights the importance of systematically analyzing the distribution of biopsy features over the entire slide and paves the way towards a personalized decision support system that will assist not only in counting cells but can also potentially improve diagnosis and provide treatment prediction.
PECNet: A Deep Multi-Label Segmentation Network for Eosinophilic Esophagitis Biopsy Diagnostics
Mar 02, 2021
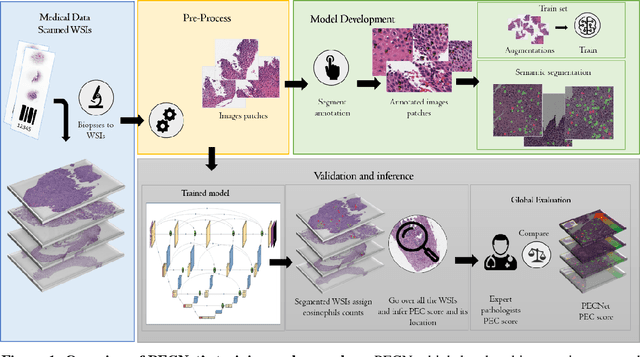
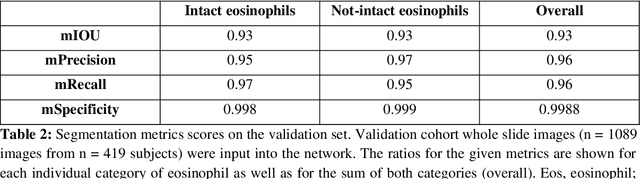

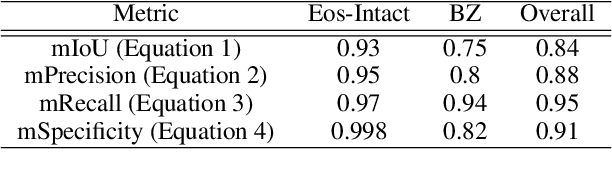
Abstract:Background. Eosinophilic esophagitis (EoE) is an allergic inflammatory condition of the esophagus associated with elevated numbers of eosinophils. Disease diagnosis and monitoring requires determining the concentration of eosinophils in esophageal biopsies, a time-consuming, tedious and somewhat subjective task currently performed by pathologists. Methods. Herein, we aimed to use machine learning to identify, quantitate and diagnose EoE. We labeled more than 100M pixels of 4345 images obtained by scanning whole slides of H&E-stained sections of esophageal biopsies derived from 23 EoE patients. We used this dataset to train a multi-label segmentation deep network. To validate the network, we examined a replication cohort of 1089 whole slide images from 419 patients derived from multiple institutions. Findings. PECNet segmented both intact and not-intact eosinophils with a mean intersection over union (mIoU) of 0.93. This segmentation was able to quantitate intact eosinophils with a mean absolute error of 0.611 eosinophils and classify EoE disease activity with an accuracy of 98.5%. Using whole slide images from the validation cohort, PECNet achieved an accuracy of 94.8%, sensitivity of 94.3%, and specificity of 95.14% in reporting EoE disease activity. Interpretation. We have developed a deep learning multi-label semantic segmentation network that successfully addresses two of the main challenges in EoE diagnostics and digital pathology, the need to detect several types of small features simultaneously and the ability to analyze whole slides efficiently. Our results pave the way for an automated diagnosis of EoE and can be utilized for other conditions with similar challenges.
Machine learning approach for biopsy-based identification of eosinophilic esophagitis reveals importance of global features
Jan 13, 2021Abstract:Goal: Eosinophilic esophagitis (EoE) is an allergic inflammatory condition characterized by eosinophil accumulation in the esophageal mucosa. EoE diagnosis includes a manual assessment of eosinophil levels in mucosal biopsies - a time-consuming, laborious task that is difficult to standardize. One of the main challenges in automating this process, like many other biopsy-based diagnostics, is detecting features that are small relative to the size of the biopsy. Results: In this work, we utilized hematoxylin- and eosin-stained slides from esophageal biopsies from patients with active EoE and control subjects to develop a platform based on a deep convolutional neural network (DCNN) that can classify esophageal biopsies with an accuracy of 85%, sensitivity of 82.5%, and specificity of 87%. Moreover, by combining several downscaling and cropping strategies, we show that some of the features contributing to the correct classification are global rather than specific, local features. Conclusions: We report the ability of artificial intelligence to identify EoE using computer vision analysis of esophageal biopsy slides. Further, the DCNN features associated with EoE are based on not only local eosinophils but also global histologic changes. Our approach can be used for other conditions that rely on biopsy-based histologic diagnostics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge