Yael Fisher
PriorPath: Coarse-To-Fine Approach for Controlled De-Novo Pathology Semantic Masks Generation
Nov 25, 2024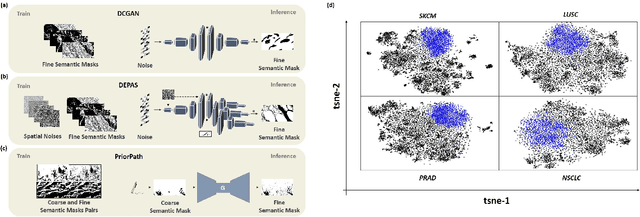
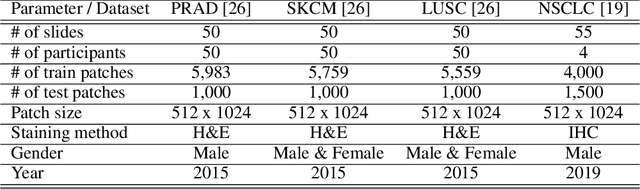
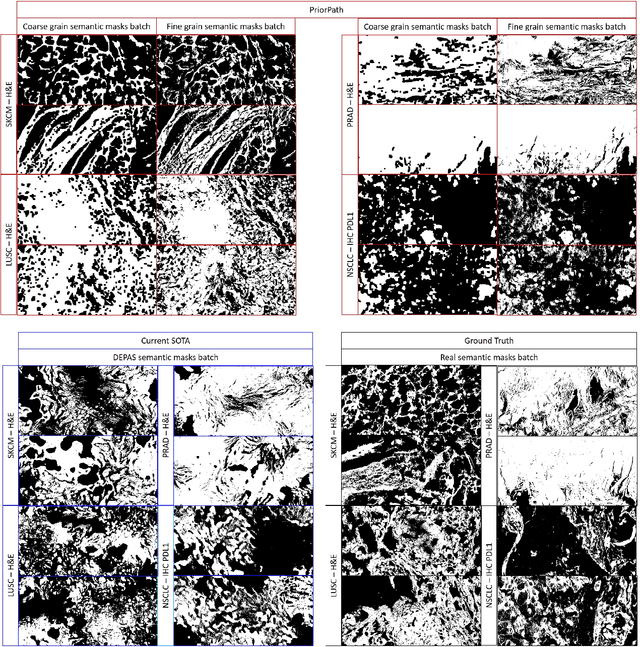
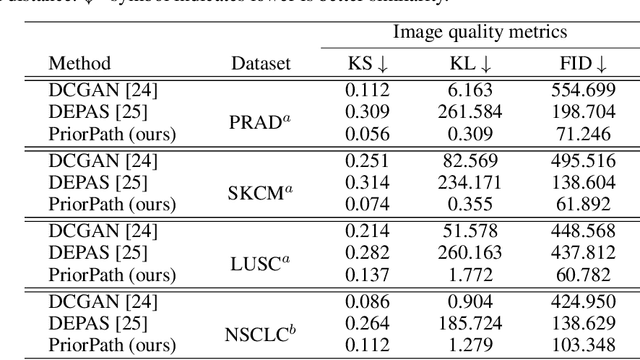
Abstract:Incorporating artificial intelligence (AI) into digital pathology offers promising prospects for automating and enhancing tasks such as image analysis and diagnostic processes. However, the diversity of tissue samples and the necessity for meticulous image labeling often result in biased datasets, constraining the applicability of algorithms trained on them. To harness synthetic histopathological images to cope with this challenge, it is essential not only to produce photorealistic images but also to be able to exert control over the cellular characteristics they depict. Previous studies used methods to generate, from random noise, semantic masks that captured the spatial distribution of the tissue. These masks were then used as a prior for conditional generative approaches to produce photorealistic histopathological images. However, as with many other generative models, this solution exhibits mode collapse as the model fails to capture the full diversity of the underlying data distribution. In this work, we present a pipeline, coined PriorPath, that generates detailed, realistic, semantic masks derived from coarse-grained images delineating tissue regions. This approach enables control over the spatial arrangement of the generated masks and, consequently, the resulting synthetic images. We demonstrated the efficacy of our method across three cancer types, skin, prostate, and lung, showcasing PriorPath's capability to cover the semantic mask space and to provide better similarity to real masks compared to previous methods. Our approach allows for specifying desired tissue distributions and obtaining both photorealistic masks and images within a single platform, thus providing a state-of-the-art, controllable solution for generating histopathological images to facilitate AI for computational pathology.
Between Generating Noise and Generating Images: Noise in the Correct Frequency Improves the Quality of Synthetic Histopathology Images for Digital Pathology
Feb 13, 2023



Abstract:Artificial intelligence and machine learning techniques have the promise to revolutionize the field of digital pathology. However, these models demand considerable amounts of data, while the availability of unbiased training data is limited. Synthetic images can augment existing datasets, to improve and validate AI algorithms. Yet, controlling the exact distribution of cellular features within them is still challenging. One of the solutions is harnessing conditional generative adversarial networks that take a semantic mask as an input rather than a random noise. Unlike other domains, outlining the exact cellular structure of tissues is hard, and most of the input masks depict regions of cell types. However, using polygon-based masks introduce inherent artifacts within the synthetic images - due to the mismatch between the polygon size and the single-cell size. In this work, we show that introducing random single-pixel noise with the appropriate spatial frequency into a polygon semantic mask can dramatically improve the quality of the synthetic images. We used our platform to generate synthetic images of immunohistochemistry-treated lung biopsies. We test the quality of the images using a three-fold validation procedure. First, we show that adding the appropriate noise frequency yields 87% of the similarity metrics improvement that is obtained by adding the actual single-cell features. Second, we show that the synthetic images pass the Turing test. Finally, we show that adding these synthetic images to the train set improves AI performance in terms of PD-L1 semantic segmentation performances. Our work suggests a simple and powerful approach for generating synthetic data on demand to unbias limited datasets to improve the algorithms' accuracy and validate their robustness.
DEPAS: De-novo Pathology Semantic Masks using a Generative Model
Feb 13, 2023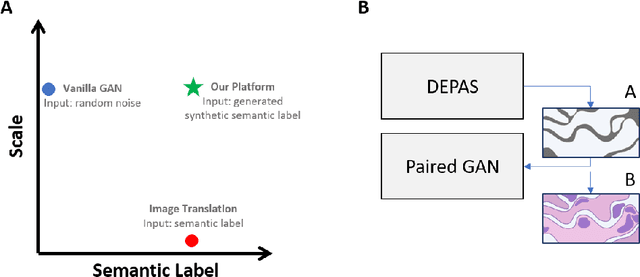
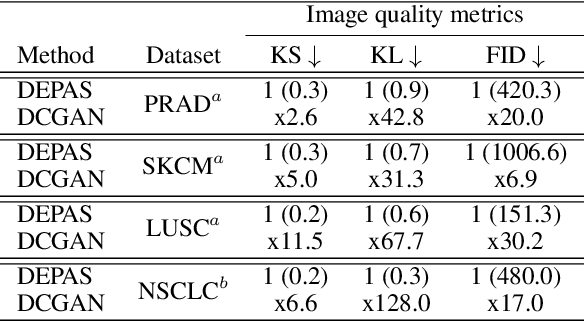
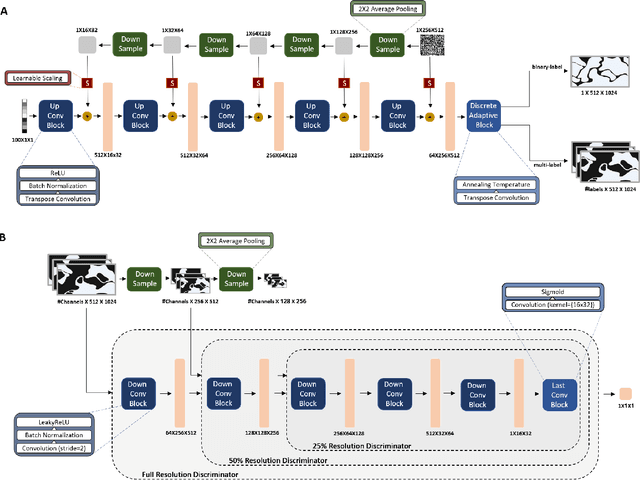
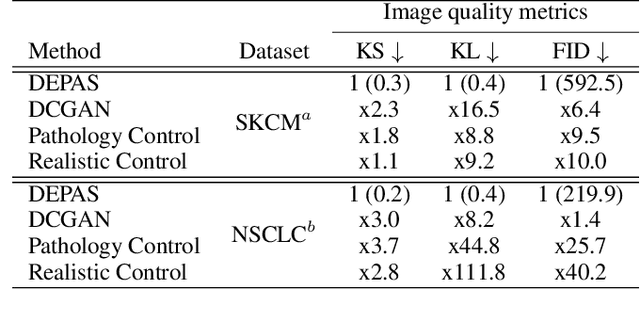
Abstract:The integration of artificial intelligence into digital pathology has the potential to automate and improve various tasks, such as image analysis and diagnostic decision-making. Yet, the inherent variability of tissues, together with the need for image labeling, lead to biased datasets that limit the generalizability of algorithms trained on them. One of the emerging solutions for this challenge is synthetic histological images. However, debiasing real datasets require not only generating photorealistic images but also the ability to control the features within them. A common approach is to use generative methods that perform image translation between semantic masks that reflect prior knowledge of the tissue and a histological image. However, unlike other image domains, the complex structure of the tissue prevents a simple creation of histology semantic masks that are required as input to the image translation model, while semantic masks extracted from real images reduce the process's scalability. In this work, we introduce a scalable generative model, coined as DEPAS, that captures tissue structure and generates high-resolution semantic masks with state-of-the-art quality. We demonstrate the ability of DEPAS to generate realistic semantic maps of tissue for three types of organs: skin, prostate, and lung. Moreover, we show that these masks can be processed using a generative image translation model to produce photorealistic histology images of two types of cancer with two different types of staining techniques. Finally, we harness DEPAS to generate multi-label semantic masks that capture different cell types distributions and use them to produce histological images with on-demand cellular features. Overall, our work provides a state-of-the-art solution for the challenging task of generating synthetic histological images while controlling their semantic information in a scalable way.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge