Muhammad Naseer Bajwa
National University of Sciences and Technology
SLUM-i: Semi-supervised Learning for Urban Mapping of Informal Settlements and Data Quality Benchmarking
Feb 04, 2026Abstract:Rapid urban expansion has fueled the growth of informal settlements in major cities of low- and middle-income countries, with Lahore and Karachi in Pakistan and Mumbai in India serving as prominent examples. However, large-scale mapping of these settlements is severely constrained not only by the scarcity of annotations but by inherent data quality challenges, specifically high spectral ambiguity between formal and informal structures and significant annotation noise. We address this by introducing a benchmark dataset for Lahore, constructed from scratch, along with companion datasets for Karachi and Mumbai, which were derived from verified administrative boundaries, totaling 1,869 $\text{km}^2$ of area. To evaluate the global robustness of our framework, we extend our experiments to five additional established benchmarks, encompassing eight cities across three continents, and provide comprehensive data quality assessments of all datasets. We also propose a new semi-supervised segmentation framework designed to mitigate the class imbalance and feature degradation inherent in standard semi-supervised learning pipelines. Our method integrates a Class-Aware Adaptive Thresholding mechanism that dynamically adjusts confidence thresholds to prevent minority class suppression and a Prototype Bank System that enforces semantic consistency by anchoring predictions to historically learned high-fidelity feature representations. Extensive experiments across a total of eight cities spanning three continents demonstrate that our approach outperforms state-of-the-art semi-supervised baselines. Most notably, our method demonstrates superior domain transfer capability whereby a model trained on only 10% of source labels reaches a 0.461 mIoU on unseen geographies and outperforms the zero-shot generalization of fully supervised models.
ExAID: A Multimodal Explanation Framework for Computer-Aided Diagnosis of Skin Lesions
Jan 04, 2022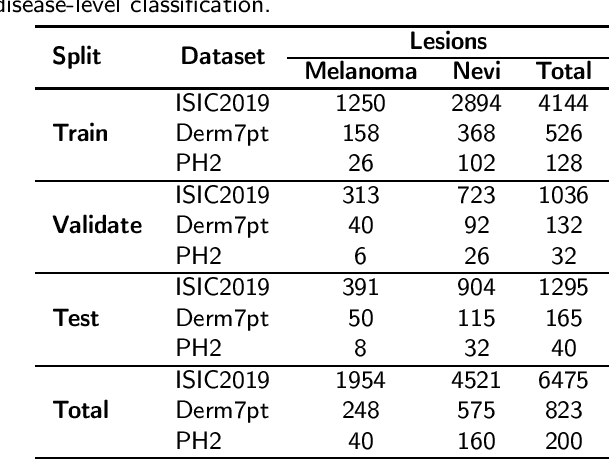
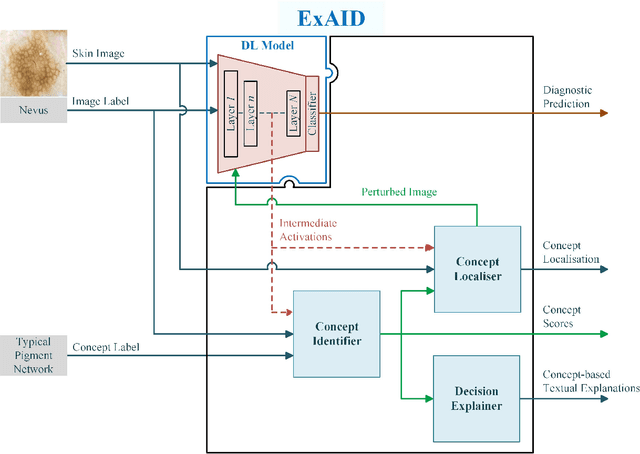
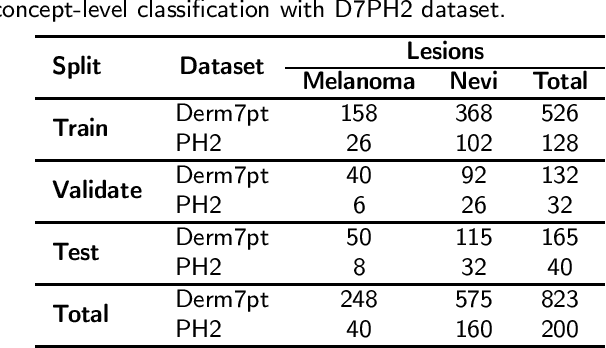
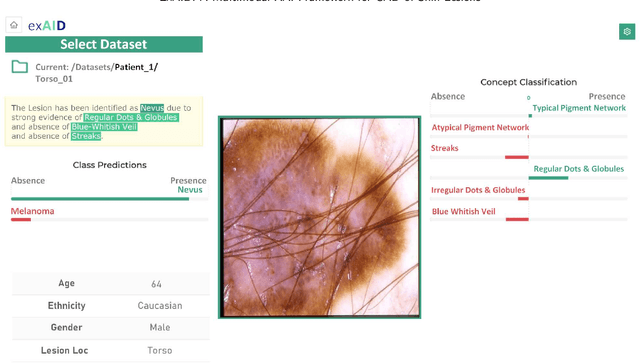
Abstract:One principal impediment in the successful deployment of AI-based Computer-Aided Diagnosis (CAD) systems in clinical workflows is their lack of transparent decision making. Although commonly used eXplainable AI methods provide some insight into opaque algorithms, such explanations are usually convoluted and not readily comprehensible except by highly trained experts. The explanation of decisions regarding the malignancy of skin lesions from dermoscopic images demands particular clarity, as the underlying medical problem definition is itself ambiguous. This work presents ExAID (Explainable AI for Dermatology), a novel framework for biomedical image analysis, providing multi-modal concept-based explanations consisting of easy-to-understand textual explanations supplemented by visual maps justifying the predictions. ExAID relies on Concept Activation Vectors to map human concepts to those learnt by arbitrary Deep Learning models in latent space, and Concept Localization Maps to highlight concepts in the input space. This identification of relevant concepts is then used to construct fine-grained textual explanations supplemented by concept-wise location information to provide comprehensive and coherent multi-modal explanations. All information is comprehensively presented in a diagnostic interface for use in clinical routines. An educational mode provides dataset-level explanation statistics and tools for data and model exploration to aid medical research and education. Through rigorous quantitative and qualitative evaluation of ExAID, we show the utility of multi-modal explanations for CAD-assisted scenarios even in case of wrong predictions. We believe that ExAID will provide dermatologists an effective screening tool that they both understand and trust. Moreover, it will be the basis for similar applications in other biomedical imaging fields.
Achievements and Challenges in Explaining Deep Learning based Computer-Aided Diagnosis Systems
Nov 26, 2020

Abstract:Remarkable success of modern image-based AI methods and the resulting interest in their applications in critical decision-making processes has led to a surge in efforts to make such intelligent systems transparent and explainable. The need for explainable AI does not stem only from ethical and moral grounds but also from stricter legislation around the world mandating clear and justifiable explanations of any decision taken or assisted by AI. Especially in the medical context where Computer-Aided Diagnosis can have a direct influence on the treatment and well-being of patients, transparency is of utmost importance for safe transition from lab research to real world clinical practice. This paper provides a comprehensive overview of current state-of-the-art in explaining and interpreting Deep Learning based algorithms in applications of medical research and diagnosis of diseases. We discuss early achievements in development of explainable AI for validation of known disease criteria, exploration of new potential biomarkers, as well as methods for the subsequent correction of AI models. Various explanation methods like visual, textual, post-hoc, ante-hoc, local and global have been thoroughly and critically analyzed. Subsequently, we also highlight some of the remaining challenges that stand in the way of practical applications of AI as a clinical decision support tool and provide recommendations for the direction of future research.
Combining Fine- and Coarse-Grained Classifiers for Diabetic Retinopathy Detection
May 28, 2020



Abstract:Visual artefacts of early diabetic retinopathy in retinal fundus images are usually small in size, inconspicuous, and scattered all over retina. Detecting diabetic retinopathy requires physicians to look at the whole image and fixate on some specific regions to locate potential biomarkers of the disease. Therefore, getting inspiration from ophthalmologist, we propose to combine coarse-grained classifiers that detect discriminating features from the whole images, with a recent breed of fine-grained classifiers that discover and pay particular attention to pathologically significant regions. To evaluate the performance of this proposed ensemble, we used publicly available EyePACS and Messidor datasets. Extensive experimentation for binary, ternary and quaternary classification shows that this ensemble largely outperforms individual image classifiers as well as most of the published works in most training setups for diabetic retinopathy detection. Furthermore, the performance of fine-grained classifiers is found notably superior than coarse-grained image classifiers encouraging the development of task-oriented fine-grained classifiers modelled after specialist ophthalmologists.
Two-stage framework for optic disc localization and glaucoma classification in retinal fundus images using deep learning
May 28, 2020



Abstract:With the advancement of powerful image processing and machine learning techniques, CAD has become ever more prevalent in all fields of medicine including ophthalmology. Since optic disc is the most important part of retinal fundus image for glaucoma detection, this paper proposes a two-stage framework that first detects and localizes optic disc and then classifies it into healthy or glaucomatous. The first stage is based on RCNN and is responsible for localizing and extracting optic disc from a retinal fundus image while the second stage uses Deep CNN to classify the extracted disc into healthy or glaucomatous. In addition to the proposed solution, we also developed a rule-based semi-automatic ground truth generation method that provides necessary annotations for training RCNN based model for automated disc localization. The proposed method is evaluated on seven publicly available datasets for disc localization and on ORIGA dataset, which is the largest publicly available dataset for glaucoma classification. The results of automatic localization mark new state-of-the-art on six datasets with accuracy reaching 100% on four of them. For glaucoma classification we achieved AUC equal to 0.874 which is 2.7% relative improvement over the state-of-the-art results previously obtained for classification on ORIGA. Once trained on carefully annotated data, Deep Learning based methods for optic disc detection and localization are not only robust, accurate and fully automated but also eliminates the need for dataset-dependent heuristic algorithms. Our empirical evaluation of glaucoma classification on ORIGA reveals that reporting only AUC, for datasets with class imbalance and without pre-defined train and test splits, does not portray true picture of the classifier's performance and calls for additional performance metrics to substantiate the results.
* 16 Pages, 10 Figures
G1020: A Benchmark Retinal Fundus Image Dataset for Computer-Aided Glaucoma Detection
May 28, 2020



Abstract:Scarcity of large publicly available retinal fundus image datasets for automated glaucoma detection has been the bottleneck for successful application of artificial intelligence towards practical Computer-Aided Diagnosis (CAD). A few small datasets that are available for research community usually suffer from impractical image capturing conditions and stringent inclusion criteria. These shortcomings in already limited choice of existing datasets make it challenging to mature a CAD system so that it can perform in real-world environment. In this paper we present a large publicly available retinal fundus image dataset for glaucoma classification called G1020. The dataset is curated by conforming to standard practices in routine ophthalmology and it is expected to serve as standard benchmark dataset for glaucoma detection. This database consists of 1020 high resolution colour fundus images and provides ground truth annotations for glaucoma diagnosis, optic disc and optic cup segmentation, vertical cup-to-disc ratio, size of neuroretinal rim in inferior, superior, nasal and temporal quadrants, and bounding box location for optic disc. We also report baseline results by conducting extensive experiments for automated glaucoma diagnosis and segmentation of optic disc and optic cup.
On Interpretability of Deep Learning based Skin Lesion Classifiers using Concept Activation Vectors
May 05, 2020
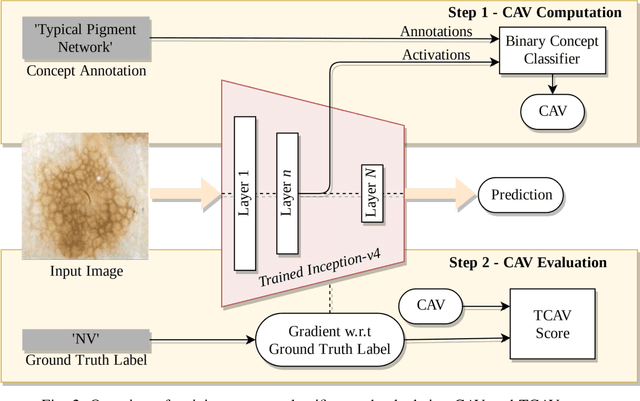

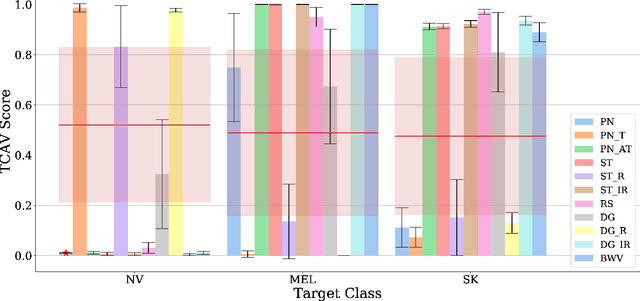
Abstract:Deep learning based medical image classifiers have shown remarkable prowess in various application areas like ophthalmology, dermatology, pathology, and radiology. However, the acceptance of these Computer-Aided Diagnosis (CAD) systems in real clinical setups is severely limited primarily because their decision-making process remains largely obscure. This work aims at elucidating a deep learning based medical image classifier by verifying that the model learns and utilizes similar disease-related concepts as described and employed by dermatologists. We used a well-trained and high performing neural network developed by REasoning for COmplex Data (RECOD) Lab for classification of three skin tumours, i.e. Melanocytic Naevi, Melanoma and Seborrheic Keratosis and performed a detailed analysis on its latent space. Two well established and publicly available skin disease datasets, PH2 and derm7pt, are used for experimentation. Human understandable concepts are mapped to RECOD image classification model with the help of Concept Activation Vectors (CAVs), introducing a novel training and significance testing paradigm for CAVs. Our results on an independent evaluation set clearly shows that the classifier learns and encodes human understandable concepts in its latent representation. Additionally, TCAV scores (Testing with CAVs) suggest that the neural network indeed makes use of disease-related concepts in the correct way when making predictions. We anticipate that this work can not only increase confidence of medical practitioners on CAD but also serve as a stepping stone for further development of CAV-based neural network interpretation methods.
Explaining AI-based Decision Support Systems using Concept Localization Maps
May 04, 2020



Abstract:Human-centric explainability of AI-based Decision Support Systems (DSS) using visual input modalities is directly related to reliability and practicality of such algorithms. An otherwise accurate and robust DSS might not enjoy trust of experts in critical application areas if it is not able to provide reasonable justification of its predictions. This paper introduces Concept Localization Maps (CLMs), which is a novel approach towards explainable image classifiers employed as DSS. CLMs extend Concept Activation Vectors (CAVs) by locating significant regions corresponding to a learned concept in the latent space of a trained image classifier. They provide qualitative and quantitative assurance of a classifier's ability to learn and focus on similar concepts important for humans during image recognition. To better understand the effectiveness of the proposed method, we generated a new synthetic dataset called Simple Concept DataBase (SCDB) that includes annotations for 10 distinguishable concepts, and made it publicly available. We evaluated our proposed method on SCDB as well as a real-world dataset called CelebA. We achieved localization recall of above 80% for most relevant concepts and average recall above 60% for all concepts using SE-ResNeXt-50 on SCDB. Our results on both datasets show great promise of CLMs for easing acceptance of DSS in practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge