Minghu Song
VEDA: 3D Molecular Generation via Variance-Exploding Diffusion with Annealing
Nov 11, 2025Abstract:Diffusion models show promise for 3D molecular generation, but face a fundamental trade-off between sampling efficiency and conformational accuracy. While flow-based models are fast, they often produce geometrically inaccurate structures, as they have difficulty capturing the multimodal distributions of molecular conformations. In contrast, denoising diffusion models are more accurate but suffer from slow sampling, a limitation attributed to sub-optimal integration between diffusion dynamics and SE(3)-equivariant architectures. To address this, we propose VEDA, a unified SE(3)-equivariant framework that combines variance-exploding diffusion with annealing to efficiently generate conformationally accurate 3D molecular structures. Specifically, our key technical contributions include: (1) a VE schedule that enables noise injection functionally analogous to simulated annealing, improving 3D accuracy and reducing relaxation energy; (2) a novel preconditioning scheme that reconciles the coordinate-predicting nature of SE(3)-equivariant networks with a residual-based diffusion objective, and (3) a new arcsin-based scheduler that concentrates sampling in critical intervals of the logarithmic signal-to-noise ratio. On the QM9 and GEOM-DRUGS datasets, VEDA matches the sampling efficiency of flow-based models, achieving state-of-the-art valency stability and validity with only 100 sampling steps. More importantly, VEDA's generated structures are remarkably stable, as measured by their relaxation energy during GFN2-xTB optimization. The median energy change is only 1.72 kcal/mol, significantly lower than the 32.3 kcal/mol from its architectural baseline, SemlaFlow. Our framework demonstrates that principled integration of VE diffusion with SE(3)-equivariant architectures can achieve both high chemical accuracy and computational efficiency.
Leveraging Partial SMILES Validation Scheme for Enhanced Drug Design in Reinforcement Learning Frameworks
May 01, 2025Abstract:SMILES-based molecule generation has emerged as a powerful approach in drug discovery. Deep reinforcement learning (RL) using large language model (LLM) has been incorporated into the molecule generation process to achieve high matching score in term of likelihood of desired molecule candidates. However, a critical challenge in this approach is catastrophic forgetting during the RL phase, where knowledge such as molecule validity, which often exceeds 99\% during pretraining, significantly deteriorates. Current RL algorithms applied in drug discovery, such as REINVENT, use prior models as anchors to retian pretraining knowledge, but these methods lack robust exploration mechanisms. To address these issues, we propose Partial SMILES Validation-PPO (PSV-PPO), a novel RL algorithm that incorporates real-time partial SMILES validation to prevent catastrophic forgetting while encouraging exploration. Unlike traditional RL approaches that validate molecule structures only after generating entire sequences, PSV-PPO performs stepwise validation at each auto-regressive step, evaluating not only the selected token candidate but also all potential branches stemming from the prior partial sequence. This enables early detection of invalid partial SMILES across all potential paths. As a result, PSV-PPO maintains high validity rates even during aggressive exploration of the vast chemical space. Our experiments on the PMO and GuacaMol benchmark datasets demonstrate that PSV-PPO significantly reduces the number of invalid generated structures while maintaining competitive exploration and optimization performance. While our work primarily focuses on maintaining validity, the framework of PSV-PPO can be extended in future research to incorporate additional forms of valuable domain knowledge, further enhancing reinforcement learning applications in drug discovery.
Binary Complex Neural Network Acceleration on FPGA
Aug 10, 2021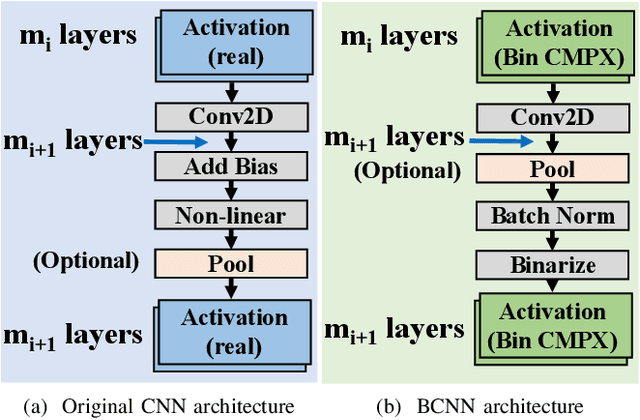
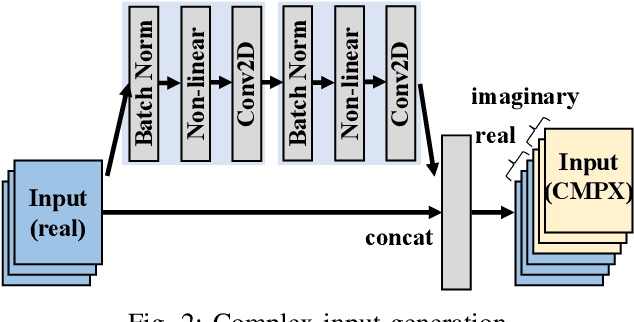
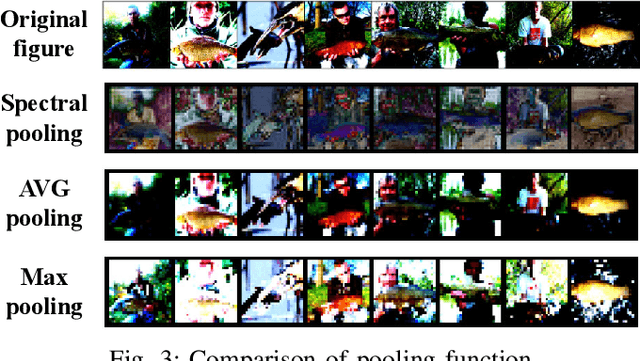
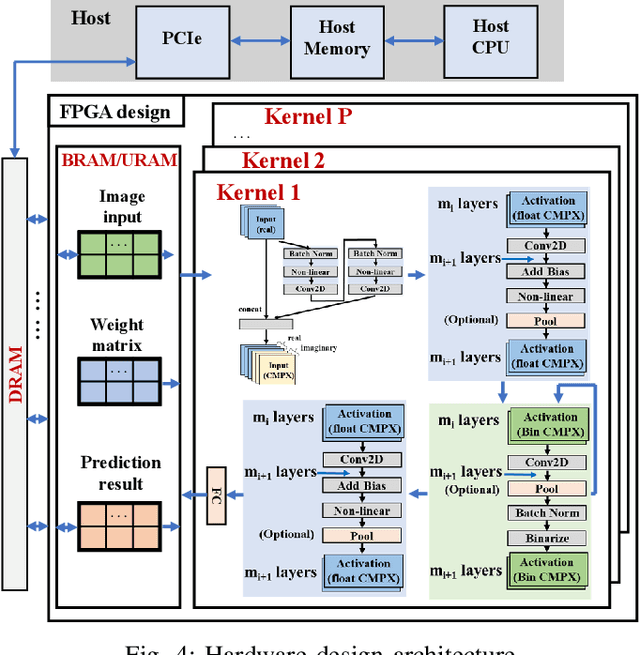
Abstract:Being able to learn from complex data with phase information is imperative for many signal processing applications. Today' s real-valued deep neural networks (DNNs) have shown efficiency in latent information analysis but fall short when applied to the complex domain. Deep complex networks (DCN), in contrast, can learn from complex data, but have high computational costs; therefore, they cannot satisfy the instant decision-making requirements of many deployable systems dealing with short observations or short signal bursts. Recent, Binarized Complex Neural Network (BCNN), which integrates DCNs with binarized neural networks (BNN), shows great potential in classifying complex data in real-time. In this paper, we propose a structural pruning based accelerator of BCNN, which is able to provide more than 5000 frames/s inference throughput on edge devices. The high performance comes from both the algorithm and hardware sides. On the algorithm side, we conduct structural pruning to the original BCNN models and obtain 20 $\times$ pruning rates with negligible accuracy loss; on the hardware side, we propose a novel 2D convolution operation accelerator for the binary complex neural network. Experimental results show that the proposed design works with over 90% utilization and is able to achieve the inference throughput of 5882 frames/s and 4938 frames/s for complex NIN-Net and ResNet-18 using CIFAR-10 dataset and Alveo U280 Board.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge