Michel Fornaciali
(De)Constructing Bias on Skin Lesion Datasets
Apr 18, 2019



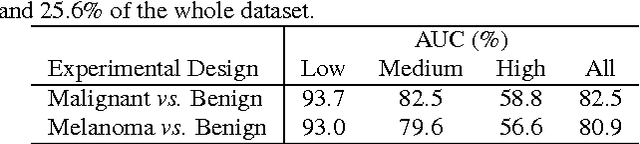
Abstract:Melanoma is the deadliest form of skin cancer. Automated skin lesion analysis plays an important role for early detection. Nowadays, the ISIC Archive and the Atlas of Dermoscopy dataset are the most employed skin lesion sources to benchmark deep-learning based tools. However, all datasets contain biases, often unintentional, due to how they were acquired and annotated. Those biases distort the performance of machine-learning models, creating spurious correlations that the models can unfairly exploit, or, contrarily destroying cogent correlations that the models could learn. In this paper, we propose a set of experiments that reveal both types of biases, positive and negative, in existing skin lesion datasets. Our results show that models can correctly classify skin lesion images without clinically-meaningful information: disturbingly, the machine-learning model learned over images where no information about the lesion remains, presents an accuracy above the AI benchmark curated with dermatologists' performances. That strongly suggests spurious correlations guiding the models. We fed models with additional clinically meaningful information, which failed to improve the results even slightly, suggesting the destruction of cogent correlations. Our main findings raise awareness of the limitations of models trained and evaluated in small datasets such as the ones we evaluated, and may suggest future guidelines for models intended for real-world deployment.
Deep-Learning Ensembles for Skin-Lesion Segmentation, Analysis, Classification: RECOD Titans at ISIC Challenge 2018
Aug 25, 2018Abstract:This extended abstract describes the participation of RECOD Titans in parts 1 to 3 of the ISIC Challenge 2018 "Skin Lesion Analysis Towards Melanoma Detection" (MICCAI 2018). Although our team has a long experience with melanoma classification and moderate experience with lesion segmentation, the ISIC Challenge 2018 was the very first time we worked on lesion attribute detection. For each task we submitted 3 different ensemble approaches, varying combinations of models and datasets. Our best results on the official testing set, regarding the official metric of each task, were: 0.728 (segmentation), 0.344 (attribute detection) and 0.803 (classification). Those submissions reached, respectively, the 56th, 14th and 9th places.
Data, Depth, and Design: Learning Reliable Models for Melanoma Screening
Mar 23, 2018



Abstract:Deep learning fostered a leap ahead in automated melanoma screening in the last two years. Those models, however, are expensive to train and difficult to parameterize. Objective: We investigate methodological issues for designing and evaluating deep learning models for melanoma detection. We explore ten choices faced by researchers: use of transfer learning, model architecture, train dataset, image resolution, type of data augmentation, input normalization, use of segmentation, duration of training, additional use of SVM, and test data augmentation. Methods: We perform two full factorial experiment, for five different test datasets, resulting in 2560 exhaustive trials in our main experiment, and 1280 trials in our assessment of transfer learning. We analyze both with multi-way ANOVA. We use the exhaustive trials to simulate sequential decisions and ensembles, with and without the use of privileged information from the test set. Results - main experiment: Amount of train data has disproportionate influence, explaining almost half the variation in performance. Of the other factors, test data augmentation and input resolution are the most influential. Deeper models, when combined, with extra data, also help. - transfer experiment: Transfer learning is critical, its absence brings huge performance penalties. - simulations: Ensembles of models are the best option to provide reliable results with limited resources, without using privileged information and sacrificing methodological rigor. Conclusions and Significance: Advancing research on automated melanoma screening requires curating larger public datasets. Indirect use of privileged information from the test set to design the models is a subtle, but frequent methodological mistake that leads to overoptimistic results. Ensembles of models are a cost-effective alternative to the expensive full-factorial and to the unstable sequential designs.
Knowledge Transfer for Melanoma Screening with Deep Learning
Mar 22, 2017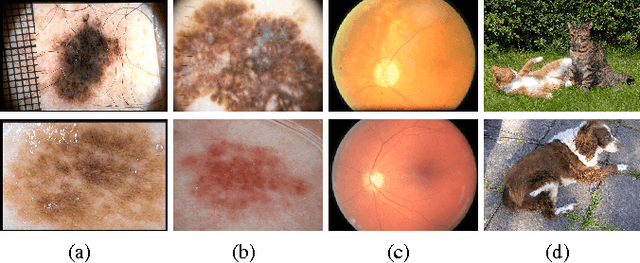


Abstract:Knowledge transfer impacts the performance of deep learning -- the state of the art for image classification tasks, including automated melanoma screening. Deep learning's greed for large amounts of training data poses a challenge for medical tasks, which we can alleviate by recycling knowledge from models trained on different tasks, in a scheme called transfer learning. Although much of the best art on automated melanoma screening employs some form of transfer learning, a systematic evaluation was missing. Here we investigate the presence of transfer, from which task the transfer is sourced, and the application of fine tuning (i.e., retraining of the deep learning model after transfer). We also test the impact of picking deeper (and more expensive) models. Our results favor deeper models, pre-trained over ImageNet, with fine-tuning, reaching an AUC of 80.7% and 84.5% for the two skin-lesion datasets evaluated.
RECOD Titans at ISIC Challenge 2017
Mar 14, 2017
Abstract:This extended abstract describes the participation of RECOD Titans in parts 1 and 3 of the ISIC Challenge 2017 "Skin Lesion Analysis Towards Melanoma Detection" (ISBI 2017). Although our team has a long experience with melanoma classification, the ISIC Challenge 2017 was the very first time we worked on skin-lesion segmentation. For part 1 (segmentation), our final submission used four of our models: two trained with all 2000 samples, without a validation split, for 250 and for 500 epochs respectively; and other two trained and validated with two different 1600/400 splits, for 220 epochs. Those four models, individually, achieved between 0.780 and 0.783 official validation scores. Our final submission averaged the output of those four models achieved a score of 0.793. For part 3 (classification), the submitted test run as well as our last official validation run were the result from a meta-model that assembled seven base deep-learning models: three based on Inception-V4 trained on our largest dataset; three based on Inception trained on our smallest dataset; and one based on ResNet-101 trained on our smaller dataset. The results of those component models were stacked in a meta-learning layer based on an SVM trained on the validation set of our largest dataset.
Towards Automated Melanoma Screening: Exploring Transfer Learning Schemes
Sep 05, 2016
Abstract:Deep learning is the current bet for image classification. Its greed for huge amounts of annotated data limits its usage in medical imaging context. In this scenario transfer learning appears as a prominent solution. In this report we aim to clarify how transfer learning schemes may influence classification results. We are particularly focused in the automated melanoma screening problem, a case of medical imaging in which transfer learning is still not widely used. We explored transfer with and without fine-tuning, sequential transfers and usage of pre-trained models in general and specific datasets. Although some issues remain open, our findings may drive future researches.
Towards Automated Melanoma Screening: Proper Computer Vision & Reliable Results
May 06, 2016



Abstract:In this paper we survey, analyze and criticize current art on automated melanoma screening, reimplementing a baseline technique, and proposing two novel ones. Melanoma, although highly curable when detected early, ends as one of the most dangerous types of cancer, due to delayed diagnosis and treatment. Its incidence is soaring, much faster than the number of trained professionals able to diagnose it. Automated screening appears as an alternative to make the most of those professionals, focusing their time on the patients at risk while safely discharging the other patients. However, the potential of automated melanoma diagnosis is currently unfulfilled, due to the emphasis of current literature on outdated computer vision models. Even more problematic is the irreproducibility of current art. We show how streamlined pipelines based upon current Computer Vision outperform conventional models - a model based on an advanced bags of words reaches an AUC of 84.6%, and a model based on deep neural networks reaches 89.3%, while the baseline (a classical bag of words) stays at 81.2%. We also initiate a dialog to improve reproducibility in our community
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge