Michal Pándy
Learning Graph Search Heuristics
Dec 07, 2022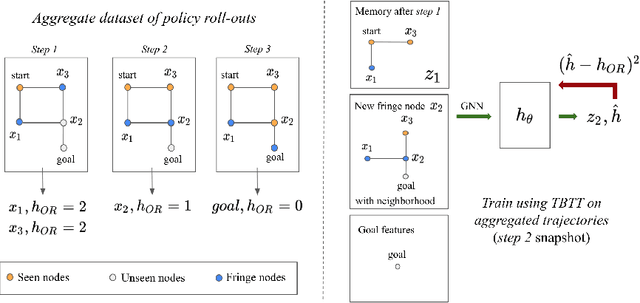
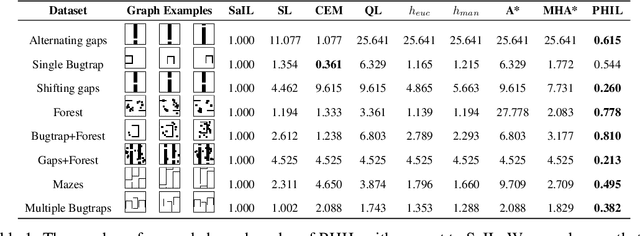
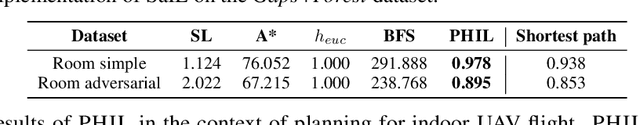
Abstract:Searching for a path between two nodes in a graph is one of the most well-studied and fundamental problems in computer science. In numerous domains such as robotics, AI, or biology, practitioners develop search heuristics to accelerate their pathfinding algorithms. However, it is a laborious and complex process to hand-design heuristics based on the problem and the structure of a given use case. Here we present PHIL (Path Heuristic with Imitation Learning), a novel neural architecture and a training algorithm for discovering graph search and navigation heuristics from data by leveraging recent advances in imitation learning and graph representation learning. At training time, we aggregate datasets of search trajectories and ground-truth shortest path distances, which we use to train a specialized graph neural network-based heuristic function using backpropagation through steps of the pathfinding process. Our heuristic function learns graph embeddings useful for inferring node distances, runs in constant time independent of graph sizes, and can be easily incorporated in an algorithm such as A* at test time. Experiments show that PHIL reduces the number of explored nodes compared to state-of-the-art methods on benchmark datasets by 58.5\% on average, can be directly applied in diverse graphs ranging from biological networks to road networks, and allows for fast planning in time-critical robotics domains.
How stable are Transferability Metrics evaluations?
Apr 11, 2022



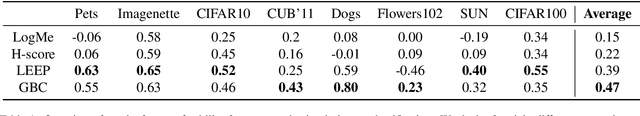
Abstract:Transferability metrics is a maturing field with increasing interest, which aims at providing heuristics for selecting the most suitable source models to transfer to a given target dataset, without fine-tuning them all. However, existing works rely on custom experimental setups which differ across papers, leading to inconsistent conclusions about which transferability metrics work best. In this paper we conduct a large-scale study by systematically constructing a broad range of 715k experimental setup variations. We discover that even small variations to an experimental setup lead to different conclusions about the superiority of a transferability metric over another. Then we propose better evaluations by aggregating across many experiments, enabling to reach more stable conclusions. As a result, we reveal the superiority of LogME at selecting good source datasets to transfer from in a semantic segmentation scenario, NLEEP at selecting good source architectures in an image classification scenario, and GBC at determining which target task benefits most from a given source model. Yet, no single transferability metric works best in all scenarios.
Transferability Estimation using Bhattacharyya Class Separability
Nov 24, 2021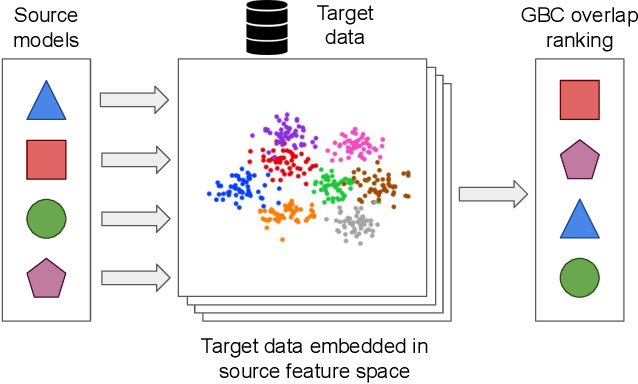


Abstract:Transfer learning has become a popular method for leveraging pre-trained models in computer vision. However, without performing computationally expensive fine-tuning, it is difficult to quantify which pre-trained source models are suitable for a specific target task, or, conversely, to which tasks a pre-trained source model can be easily adapted to. In this work, we propose Gaussian Bhattacharyya Coefficient (GBC), a novel method for quantifying transferability between a source model and a target dataset. In a first step we embed all target images in the feature space defined by the source model, and represent them with per-class Gaussians. Then, we estimate their pairwise class separability using the Bhattacharyya coefficient, yielding a simple and effective measure of how well the source model transfers to the target task. We evaluate GBC on image classification tasks in the context of dataset and architecture selection. Further, we also perform experiments on the more complex semantic segmentation transferability estimation task. We demonstrate that GBC outperforms state-of-the-art transferability metrics on most evaluation criteria in the semantic segmentation settings, matches the performance of top methods for dataset transferability in image classification, and performs best on architecture selection problems for image classification.
Neural Distance Embeddings for Biological Sequences
Oct 11, 2021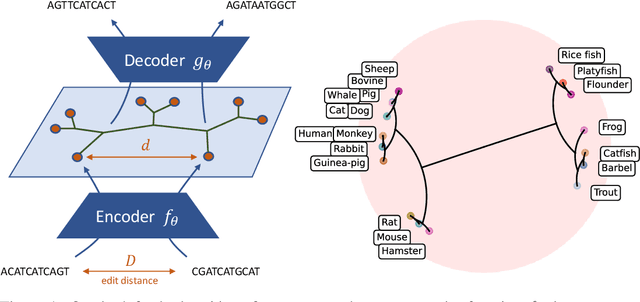


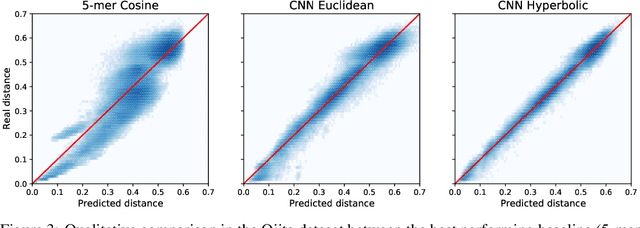
Abstract:The development of data-dependent heuristics and representations for biological sequences that reflect their evolutionary distance is critical for large-scale biological research. However, popular machine learning approaches, based on continuous Euclidean spaces, have struggled with the discrete combinatorial formulation of the edit distance that models evolution and the hierarchical relationship that characterises real-world datasets. We present Neural Distance Embeddings (NeuroSEED), a general framework to embed sequences in geometric vector spaces, and illustrate the effectiveness of the hyperbolic space that captures the hierarchical structure and provides an average 22% reduction in embedding RMSE against the best competing geometry. The capacity of the framework and the significance of these improvements are then demonstrated devising supervised and unsupervised NeuroSEED approaches to multiple core tasks in bioinformatics. Benchmarked with common baselines, the proposed approaches display significant accuracy and/or runtime improvements on real-world datasets. As an example for hierarchical clustering, the proposed pretrained and from-scratch methods match the quality of competing baselines with 30x and 15x runtime reduction, respectively.
Learning To Find Shortest Collision-Free Paths From Images
Nov 30, 2020



Abstract:Motion planning is a fundamental problem in robotics and machine perception. Sampling-based planners find accurate solutions by exhaustively exploring the space, but are inefficient and tend to produce jerky motions. Optimization and learning-based planners are more efficient and produce smooth trajectories. However, a significant hurdle that these approaches face is constructing a differentiable cost function that simultaneously minimizes path length and avoids collisions. These two objectives are conflicting by nature -- path length is continuous and well-behaved, but collisions are discrete non-differentiable events. Reconciling these terms has been a significant challenge in optimization-based motion planning. The main contribution of this paper is a novel cost function that guarantees collision-free shortest paths are found at its minimum. We show that our approach works seamlessly with RGBD input and predicts high-quality paths in 2D, 3D, and 6 DoF robotic manipulator settings. Our method also reduces training and inference time compared to existing approaches, in some cases by orders of magnitude.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge