Michael Morris
University of Maryland, Baltimore County, National Institutes of Health Clinical Center, Networking Health
Forecasting infectious disease prevalence with associated uncertainty using neural networks
Sep 02, 2024
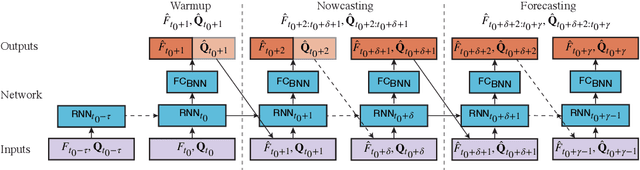
Abstract:Infectious diseases pose significant human and economic burdens. Accurately forecasting disease incidence can enable public health agencies to respond effectively to existing or emerging diseases. Despite progress in the field, developing accurate forecasting models remains a significant challenge. This thesis proposes two methodological frameworks using neural networks (NNs) with associated uncertainty estimates - a critical component limiting the application of NNs to epidemic forecasting thus far. We develop our frameworks by forecasting influenza-like illness (ILI) in the United States. Our first proposed method uses Web search activity data in conjunction with historical ILI rates as observations for training NN architectures. Our models incorporate Bayesian layers to produce uncertainty intervals, positioning themselves as legitimate alternatives to more conventional approaches. The best performing architecture: iterative recurrent neural network (IRNN), reduces mean absolute error by 10.3% and improves Skill by 17.1% on average in forecasting tasks across four flu seasons compared to the state-of-the-art. We build on this method by introducing IRNNs, an architecture which changes the sampling procedure in the IRNN to improve the uncertainty estimation. Our second framework uses neural ordinary differential equations to bridge the gap between mechanistic compartmental models and NNs; benefiting from the physical constraints that compartmental models provide. We evaluate eight neural ODE models utilising a mixture of ILI rates and Web search activity data to provide forecasts. These are compared with the IRNN and IRNN0 - the IRNN using only ILI rates. Models trained without Web search activity data outperform the IRNN0 by 16% in terms of Skill. Future work should focus on more effectively using neural ODEs with Web search data to compete with the best performing IRNN.
The Impact of an XAI-Augmented Approach on Binary Classification with Scarce Data
Jul 01, 2024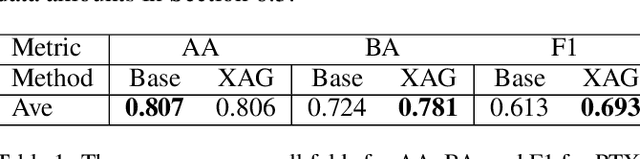
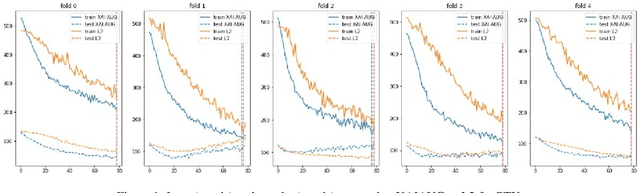
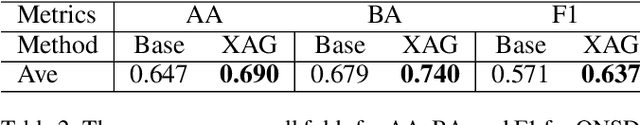
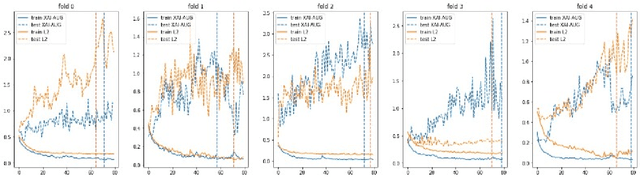
Abstract:Point-of-Care Ultrasound (POCUS) is the practice of clinicians conducting and interpreting ultrasound scans right at the patient's bedside. However, the expertise needed to interpret these images is considerable and may not always be present in emergency situations. This reality makes algorithms such as machine learning classifiers extremely valuable to augment human decisions. POCUS devices are becoming available at a reasonable cost in the size of a mobile phone. The challenge of turning POCUS devices into life-saving tools is that interpretation of ultrasound images requires specialist training and experience. Unfortunately, the difficulty to obtain positive training images represents an important obstacle to building efficient and accurate classifiers. Hence, the problem we try to investigate is how to explore strategies to increase accuracy of classifiers trained with scarce data. We hypothesize that training with a few data instances may not suffice for classifiers to generalize causing them to overfit. Our approach uses an Explainable AI-Augmented approach to help the algorithm learn more from less and potentially help the classifier better generalize.
CCS-GAN: COVID-19 CT-scan classification with very few positive training images
Oct 01, 2021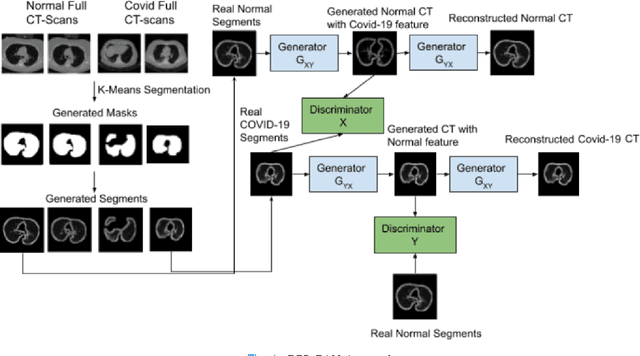

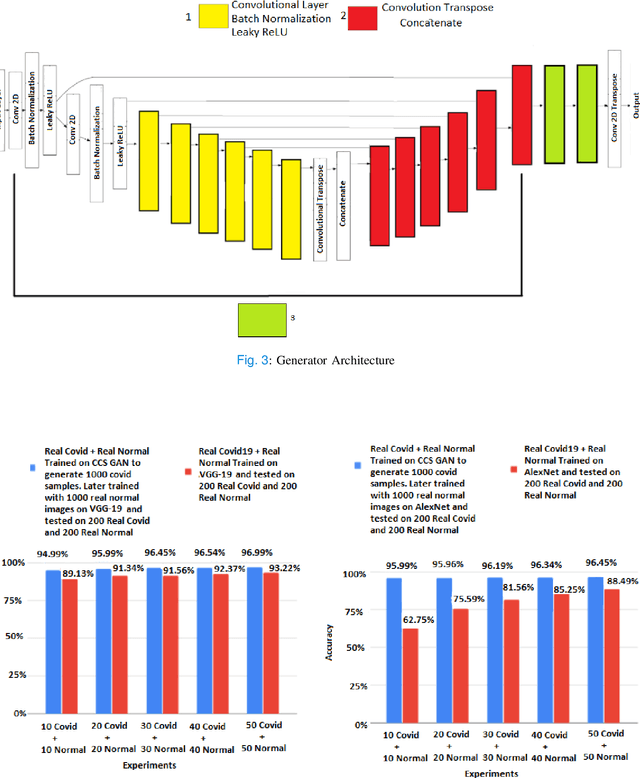
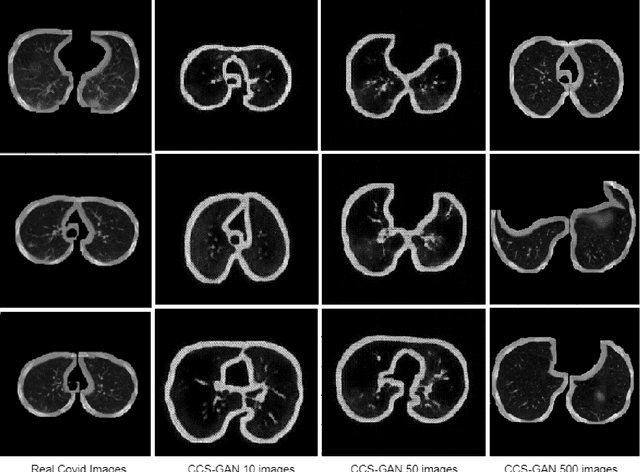
Abstract:We present a novel algorithm that is able to classify COVID-19 pneumonia from CT Scan slices using a very small sample of training images exhibiting COVID-19 pneumonia in tandem with a larger number of normal images. This algorithm is able to achieve high classification accuracy using as few as 10 positive training slices (from 10 positive cases), which to the best of our knowledge is one order of magnitude fewer than the next closest published work at the time of writing. Deep learning with extremely small positive training volumes is a very difficult problem and has been an important topic during the COVID-19 pandemic, because for quite some time it was difficult to obtain large volumes of COVID-19 positive images for training. Algorithms that can learn to screen for diseases using few examples are an important area of research. We present the Cycle Consistent Segmentation Generative Adversarial Network (CCS-GAN). CCS-GAN combines style transfer with pulmonary segmentation and relevant transfer learning from negative images in order to create a larger volume of synthetic positive images for the purposes of improving diagnostic classification performance. The performance of a VGG-19 classifier plus CCS-GAN was trained using a small sample of positive image slices ranging from at most 50 down to as few as 10 COVID-19 positive CT-scan images. CCS-GAN achieves high accuracy with few positive images and thereby greatly reduces the barrier of acquiring large training volumes in order to train a diagnostic classifier for COVID-19.
Estimating the Uncertainty of Neural Network Forecasts for Influenza Prevalence Using Web Search Activity
May 26, 2021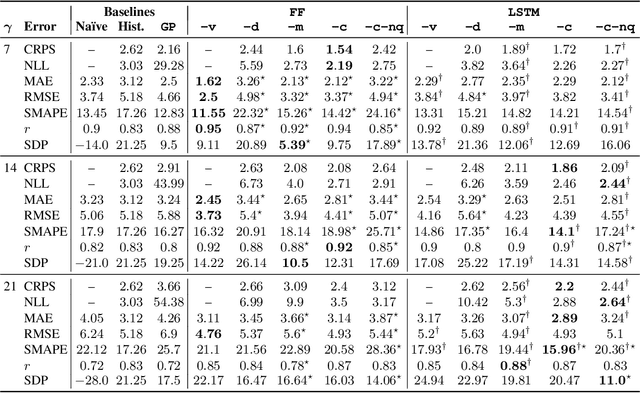
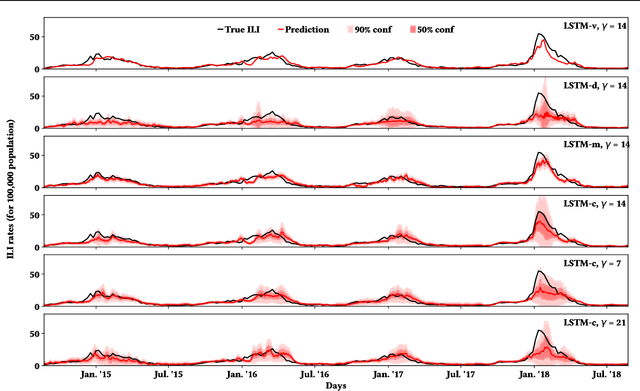
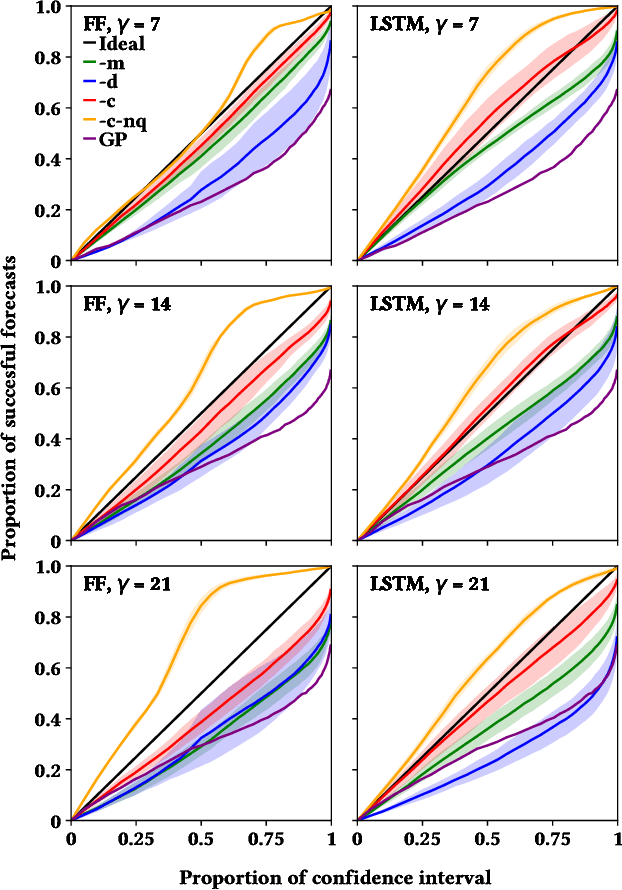
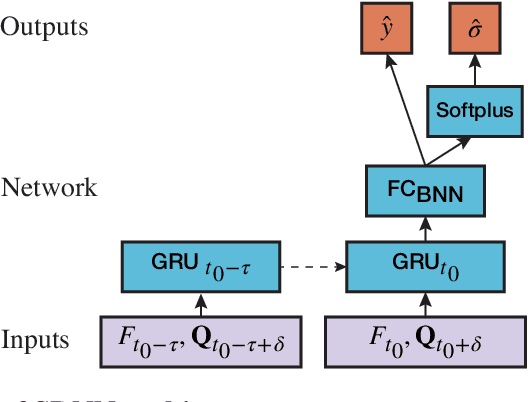
Abstract:Influenza is an infectious disease with the potential to become a pandemic, and hence, forecasting its prevalence is an important undertaking for planning an effective response. Research has found that web search activity can be used to improve influenza models. Neural networks (NN) can provide state-of-the-art forecasting accuracy but do not commonly incorporate uncertainty in their estimates, something essential for using them effectively during decision making. In this paper, we demonstrate how Bayesian Neural Networks (BNNs) can be used to both provide a forecast and a corresponding uncertainty without significant loss in forecasting accuracy compared to traditional NNs. Our method accounts for two sources of uncertainty: data and model uncertainty, arising due to measurement noise and model specification, respectively. Experiments are conducted using 14 years of data for England, assessing the model's accuracy over the last 4 flu seasons in this dataset. We evaluate the performance of different models including competitive baselines with conventional metrics as well as error functions that incorporate uncertainty estimates. Our empirical analysis indicates that considering both sources of uncertainty simultaneously is superior to considering either one separately. We also show that a BNN with recurrent layers that models both sources of uncertainty yields superior accuracy for these metrics for forecasting horizons greater than 7 days.
Toward Generating Synthetic CT Volumes using a 3D-Conditional Generative Adversarial Network
Apr 02, 2021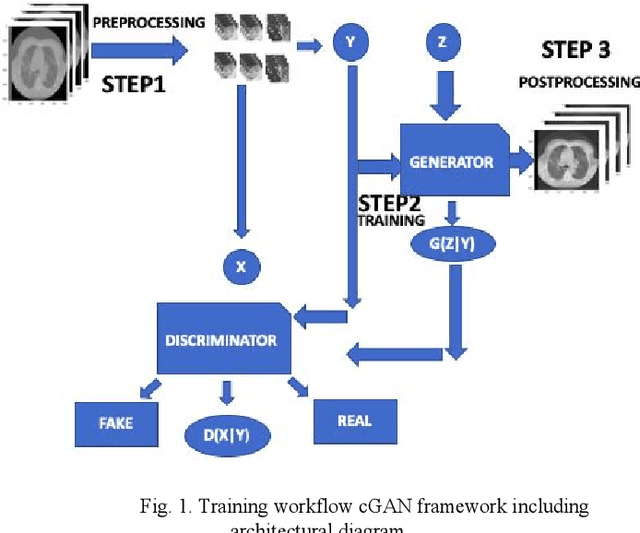
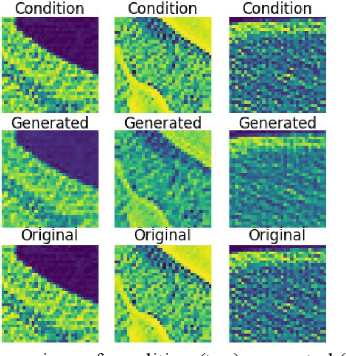
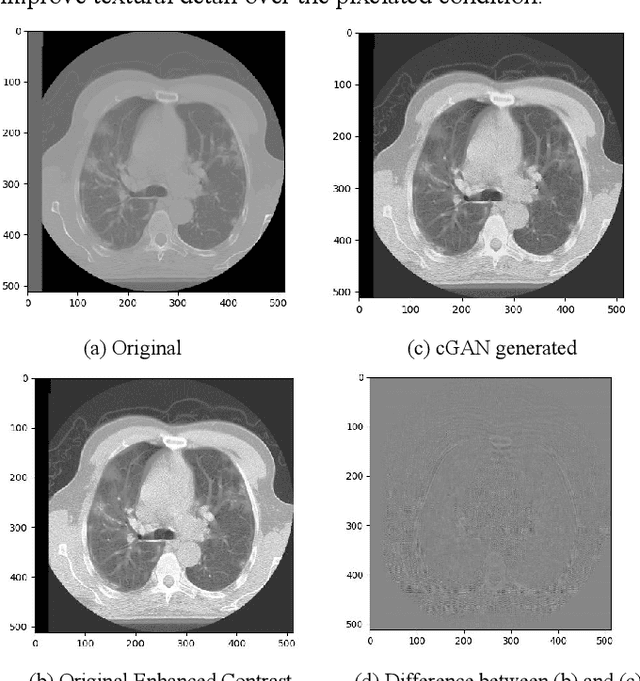
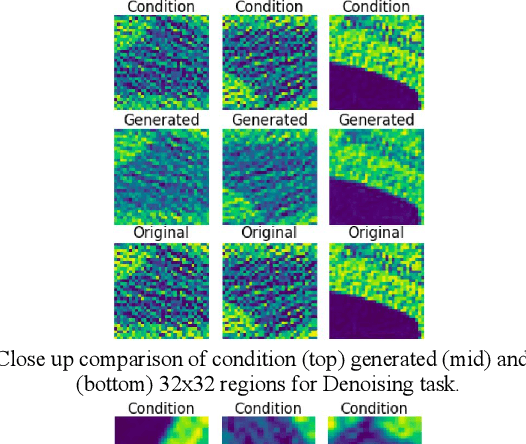
Abstract:We present a novel conditional Generative Adversarial Network (cGAN) architecture that is capable of generating 3D Computed Tomography scans in voxels from noisy and/or pixelated approximations and with the potential to generate full synthetic 3D scan volumes. We believe conditional cGAN to be a tractable approach to generate 3D CT volumes, even though the problem of generating full resolution deep fakes is presently impractical due to GPU memory limitations. We present results for autoencoder, denoising, and depixelating tasks which are trained and tested on two novel COVID19 CT datasets. Our evaluation metrics, Peak Signal to Noise ratio (PSNR) range from 12.53 - 46.46 dB, and the Structural Similarity index ( SSIM) range from 0.89 to 1.
Deep Expectation-Maximization for Semi-Supervised Lung Cancer Screening
Oct 02, 2020
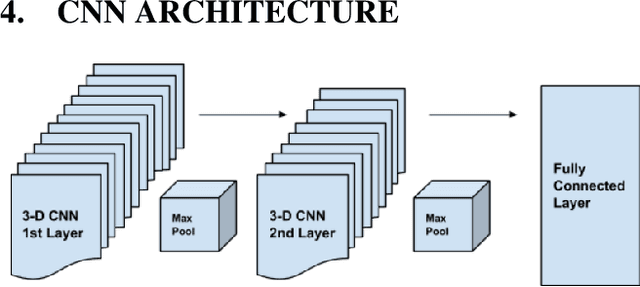
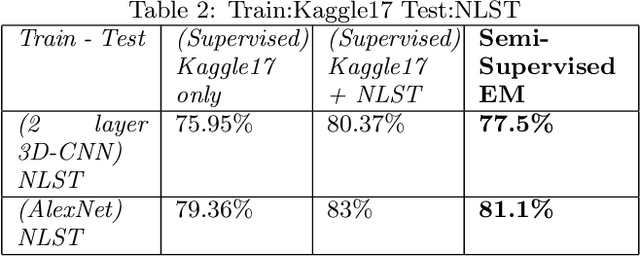
Abstract:We present a semi-supervised algorithm for lung cancer screening in which a 3D Convolutional Neural Network (CNN) is trained using the Expectation-Maximization (EM) meta-algorithm. Semi-supervised learning allows a smaller labelled data-set to be combined with an unlabeled data-set in order to provide a larger and more diverse training sample. EM allows the algorithm to simultaneously calculate a maximum likelihood estimate of the CNN training coefficients along with the labels for the unlabeled training set which are defined as a latent variable space. We evaluate the model performance of the Semi-Supervised EM algorithm for CNNs through cross-domain training of the Kaggle Data Science Bowl 2017 (Kaggle17) data-set with the National Lung Screening Trial (NLST) data-set. Our results show that the Semi-Supervised EM algorithm greatly improves the classification accuracy of the cross-domain lung cancer screening, although results are lower than a fully supervised approach with the advantage of additional labelled data from the unsupervised sample. As such, we demonstrate that Semi-Supervised EM is a valuable technique to improve the accuracy of lung cancer screening models using 3D CNNs.
Generating Realistic COVID19 X-rays with a Mean Teacher + Transfer Learning GAN
Sep 26, 2020
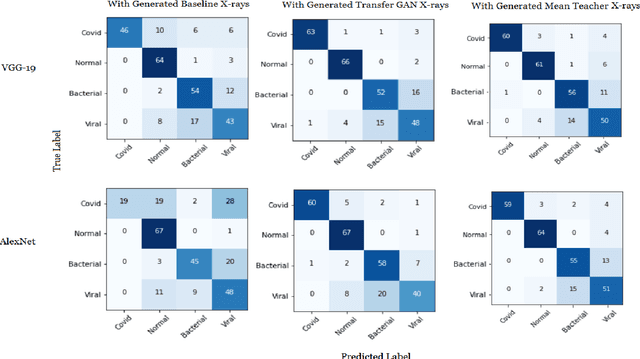
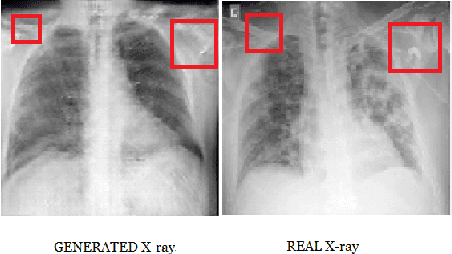
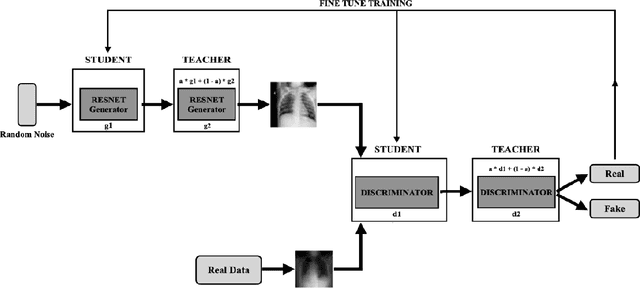
Abstract:COVID-19 is a novel infectious disease responsible for over 800K deaths worldwide as of August 2020. The need for rapid testing is a high priority and alternative testing strategies including X-ray image classification are a promising area of research. However, at present, public datasets for COVID19 x-ray images have low data volumes, making it challenging to develop accurate image classifiers. Several recent papers have made use of Generative Adversarial Networks (GANs) in order to increase the training data volumes. But realistic synthetic COVID19 X-rays remain challenging to generate. We present a novel Mean Teacher + Transfer GAN (MTT-GAN) that generates COVID19 chest X-ray images of high quality. In order to create a more accurate GAN, we employ transfer learning from the Kaggle Pneumonia X-Ray dataset, a highly relevant data source orders of magnitude larger than public COVID19 datasets. Furthermore, we employ the Mean Teacher algorithm as a constraint to improve stability of training. Our qualitative analysis shows that the MTT-GAN generates X-ray images that are greatly superior to a baseline GAN and visually comparable to real X-rays. Although board-certified radiologists can distinguish MTT-GAN fakes from real COVID19 X-rays. Quantitative analysis shows that MTT-GAN greatly improves the accuracy of both a binary COVID19 classifier as well as a multi-class Pneumonia classifier as compared to a baseline GAN. Our classification accuracy is favourable as compared to recently reported results in the literature for similar binary and multi-class COVID19 screening tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge