Sumeet Menon
University of Maryland, Baltimore County
A Method of Moments Embedding Constraint and its Application to Semi-Supervised Learning
Apr 27, 2024Abstract:Discriminative deep learning models with a linear+softmax final layer have a problem: the latent space only predicts the conditional probabilities $p(Y|X)$ but not the full joint distribution $p(Y,X)$, which necessitates a generative approach. The conditional probability cannot detect outliers, causing outlier sensitivity in softmax networks. This exacerbates model over-confidence impacting many problems, such as hallucinations, confounding biases, and dependence on large datasets. To address this we introduce a novel embedding constraint based on the Method of Moments (MoM). We investigate the use of polynomial moments ranging from 1st through 4th order hyper-covariance matrices. Furthermore, we use this embedding constraint to train an Axis-Aligned Gaussian Mixture Model (AAGMM) final layer, which learns not only the conditional, but also the joint distribution of the latent space. We apply this method to the domain of semi-supervised image classification by extending FlexMatch with our technique. We find our MoM constraint with the AAGMM layer is able to match the reported FlexMatch accuracy, while also modeling the joint distribution, thereby reducing outlier sensitivity. We also present a preliminary outlier detection strategy based on Mahalanobis distance and discuss future improvements to this strategy. Code is available at: \url{https://github.com/mmajurski/ssl-gmm}
Semi-supervised Contrastive Outlier removal for Pseudo Expectation Maximization (SCOPE)
Jun 28, 2022



Abstract:Semi-supervised learning is the problem of training an accurate predictive model by combining a small labeled dataset with a presumably much larger unlabeled dataset. Many methods for semi-supervised deep learning have been developed, including pseudolabeling, consistency regularization, and contrastive learning techniques. Pseudolabeling methods however are highly susceptible to confounding, in which erroneous pseudolabels are assumed to be true labels in early iterations, thereby causing the model to reinforce its prior biases and thereby fail to generalize to strong predictive performance. We present a new approach to suppress confounding errors through a method we describe as Semi-supervised Contrastive Outlier removal for Pseudo Expectation Maximization (SCOPE). Like basic pseudolabeling, SCOPE is related to Expectation Maximization (EM), a latent variable framework which can be extended toward understanding cluster-assumption deep semi-supervised algorithms. However, unlike basic pseudolabeling which fails to adequately take into account the probability of the unlabeled samples given the model, SCOPE introduces an outlier suppression term designed to improve the behavior of EM iteration given a discrimination DNN backbone in the presence of outliers. Our results show that SCOPE greatly improves semi-supervised classification accuracy over a baseline, and furthermore when combined with consistency regularization achieves the highest reported accuracy for the semi-supervised CIFAR-10 classification task using 250 and 4000 labeled samples. Moreover, we show that SCOPE reduces the prevalence of confounding errors during pseudolabeling iterations by pruning erroneous high-confidence pseudolabeled samples that would otherwise contaminate the labeled set in subsequent retraining iterations.
CCS-GAN: COVID-19 CT-scan classification with very few positive training images
Oct 01, 2021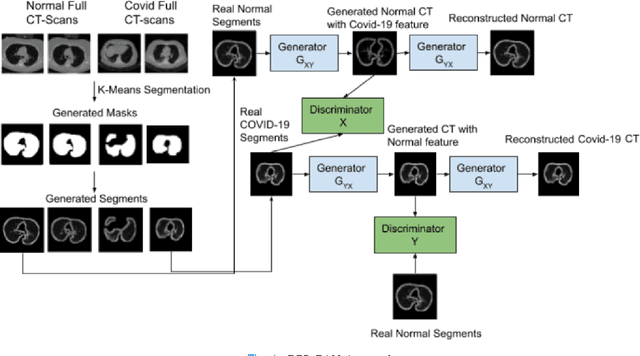

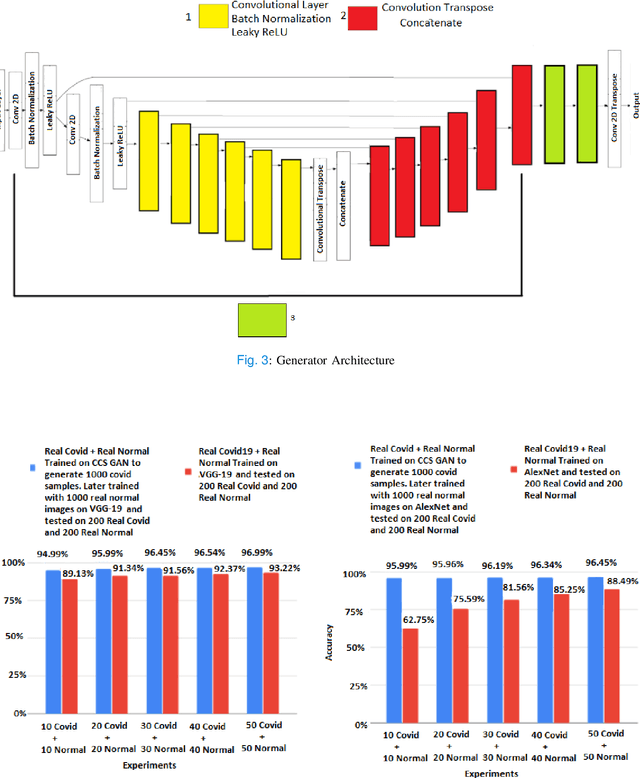
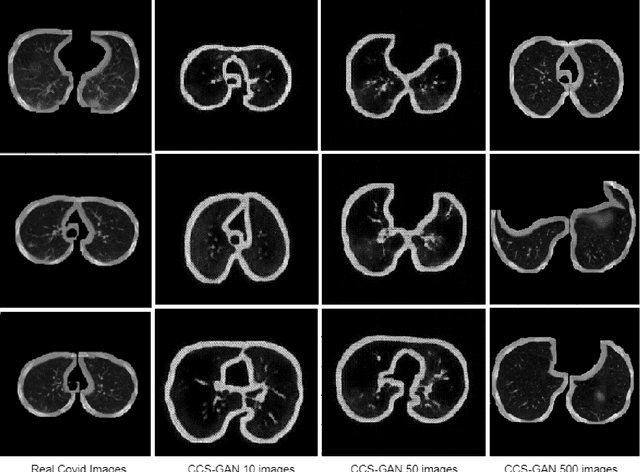
Abstract:We present a novel algorithm that is able to classify COVID-19 pneumonia from CT Scan slices using a very small sample of training images exhibiting COVID-19 pneumonia in tandem with a larger number of normal images. This algorithm is able to achieve high classification accuracy using as few as 10 positive training slices (from 10 positive cases), which to the best of our knowledge is one order of magnitude fewer than the next closest published work at the time of writing. Deep learning with extremely small positive training volumes is a very difficult problem and has been an important topic during the COVID-19 pandemic, because for quite some time it was difficult to obtain large volumes of COVID-19 positive images for training. Algorithms that can learn to screen for diseases using few examples are an important area of research. We present the Cycle Consistent Segmentation Generative Adversarial Network (CCS-GAN). CCS-GAN combines style transfer with pulmonary segmentation and relevant transfer learning from negative images in order to create a larger volume of synthetic positive images for the purposes of improving diagnostic classification performance. The performance of a VGG-19 classifier plus CCS-GAN was trained using a small sample of positive image slices ranging from at most 50 down to as few as 10 COVID-19 positive CT-scan images. CCS-GAN achieves high accuracy with few positive images and thereby greatly reduces the barrier of acquiring large training volumes in order to train a diagnostic classifier for COVID-19.
Toward Generating Synthetic CT Volumes using a 3D-Conditional Generative Adversarial Network
Apr 02, 2021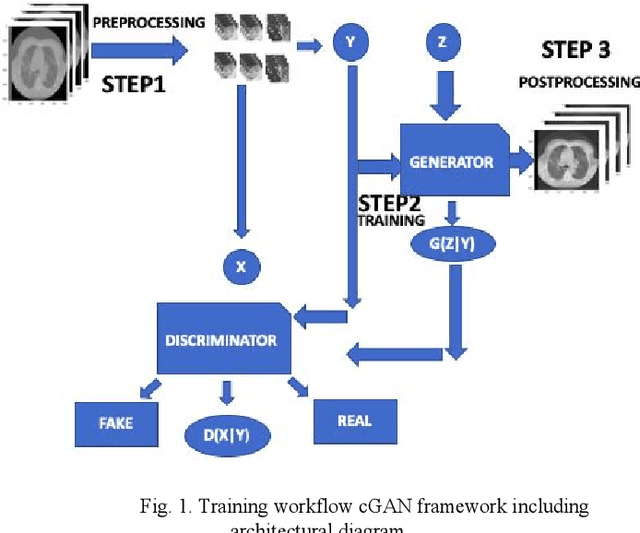
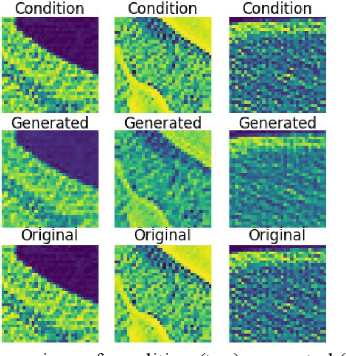
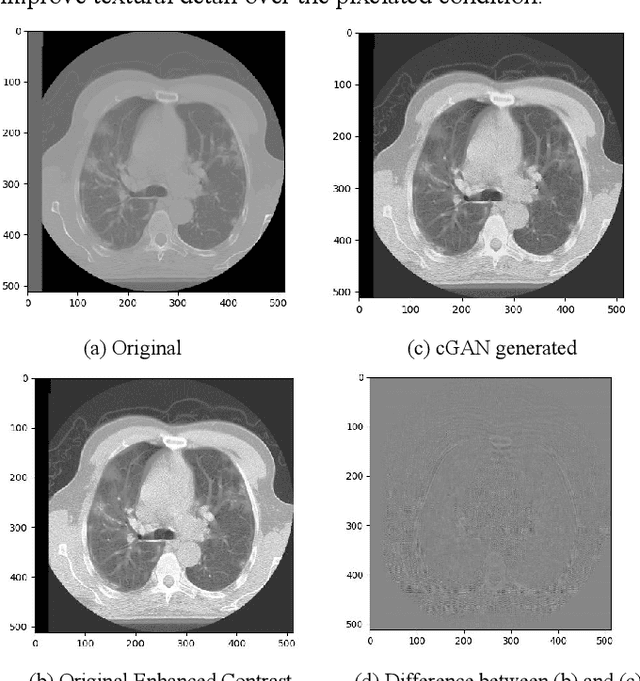
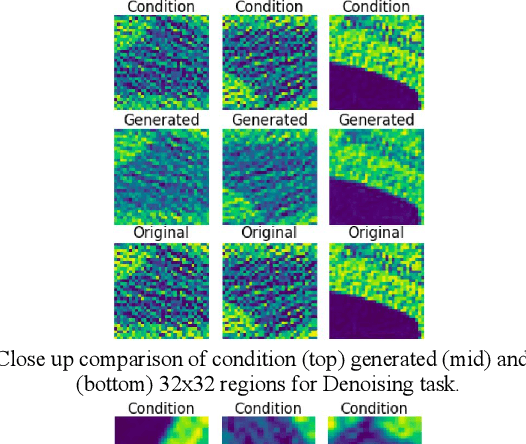
Abstract:We present a novel conditional Generative Adversarial Network (cGAN) architecture that is capable of generating 3D Computed Tomography scans in voxels from noisy and/or pixelated approximations and with the potential to generate full synthetic 3D scan volumes. We believe conditional cGAN to be a tractable approach to generate 3D CT volumes, even though the problem of generating full resolution deep fakes is presently impractical due to GPU memory limitations. We present results for autoencoder, denoising, and depixelating tasks which are trained and tested on two novel COVID19 CT datasets. Our evaluation metrics, Peak Signal to Noise ratio (PSNR) range from 12.53 - 46.46 dB, and the Structural Similarity index ( SSIM) range from 0.89 to 1.
Lung Nodule Classification Using Biomarkers, Volumetric Radiomics and 3D CNNs
Oct 19, 2020



Abstract:We present a hybrid algorithm to estimate lung nodule malignancy that combines imaging biomarkers from Radiologist's annotation with image classification of CT scans. Our algorithm employs a 3D Convolutional Neural Network (CNN) as well as a Random Forest in order to combine CT imagery with biomarker annotation and volumetric radiomic features. We analyze and compare the performance of the algorithm using only imagery, only biomarkers, combined imagery + biomarkers, combined imagery + volumetric radiomic features and finally the combination of imagery + biomarkers + volumetric features in order to classify the suspicion level of nodule malignancy. The National Cancer Institute (NCI) Lung Image Database Consortium (LIDC) IDRI dataset is used to train and evaluate the classification task. We show that the incorporation of semi-supervised learning by means of K-Nearest-Neighbors (KNN) can increase the available training sample size of the LIDC-IDRI thereby further improving the accuracy of malignancy estimation of most of the models tested although there is no significant improvement with the use of KNN semi-supervised learning if image classification with CNNs and volumetric features are combined with descriptive biomarkers. Unexpectedly, we also show that a model using image biomarkers alone is more accurate than one that combines biomarkers with volumetric radiomics, 3D CNNs, and semi-supervised learning. We discuss the possibility that this result may be influenced by cognitive bias in LIDC-IDRI because malignancy estimates were recorded by the same radiologist panel as biomarkers, as well as future work to incorporate pathology information over a subset of study participants.
Deep Expectation-Maximization for Semi-Supervised Lung Cancer Screening
Oct 02, 2020
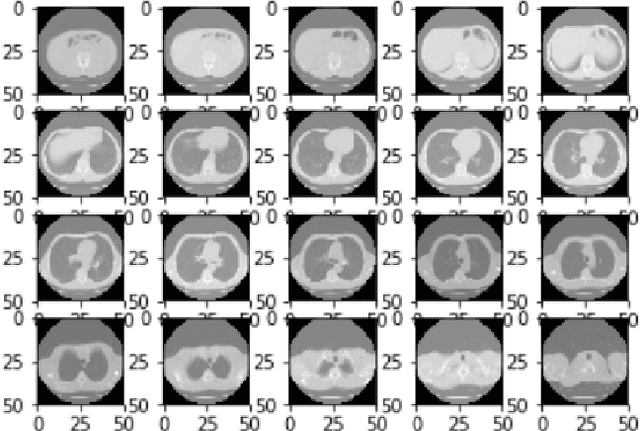
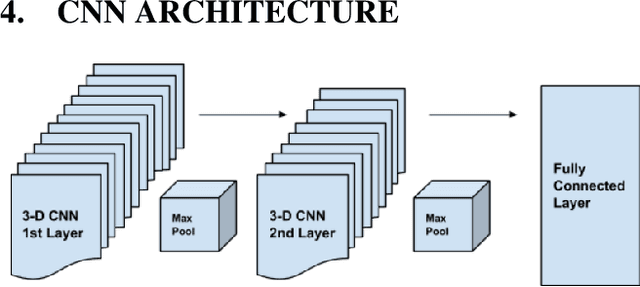
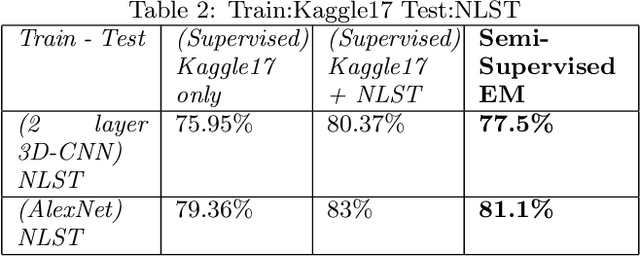
Abstract:We present a semi-supervised algorithm for lung cancer screening in which a 3D Convolutional Neural Network (CNN) is trained using the Expectation-Maximization (EM) meta-algorithm. Semi-supervised learning allows a smaller labelled data-set to be combined with an unlabeled data-set in order to provide a larger and more diverse training sample. EM allows the algorithm to simultaneously calculate a maximum likelihood estimate of the CNN training coefficients along with the labels for the unlabeled training set which are defined as a latent variable space. We evaluate the model performance of the Semi-Supervised EM algorithm for CNNs through cross-domain training of the Kaggle Data Science Bowl 2017 (Kaggle17) data-set with the National Lung Screening Trial (NLST) data-set. Our results show that the Semi-Supervised EM algorithm greatly improves the classification accuracy of the cross-domain lung cancer screening, although results are lower than a fully supervised approach with the advantage of additional labelled data from the unsupervised sample. As such, we demonstrate that Semi-Supervised EM is a valuable technique to improve the accuracy of lung cancer screening models using 3D CNNs.
Generating Realistic COVID19 X-rays with a Mean Teacher + Transfer Learning GAN
Sep 26, 2020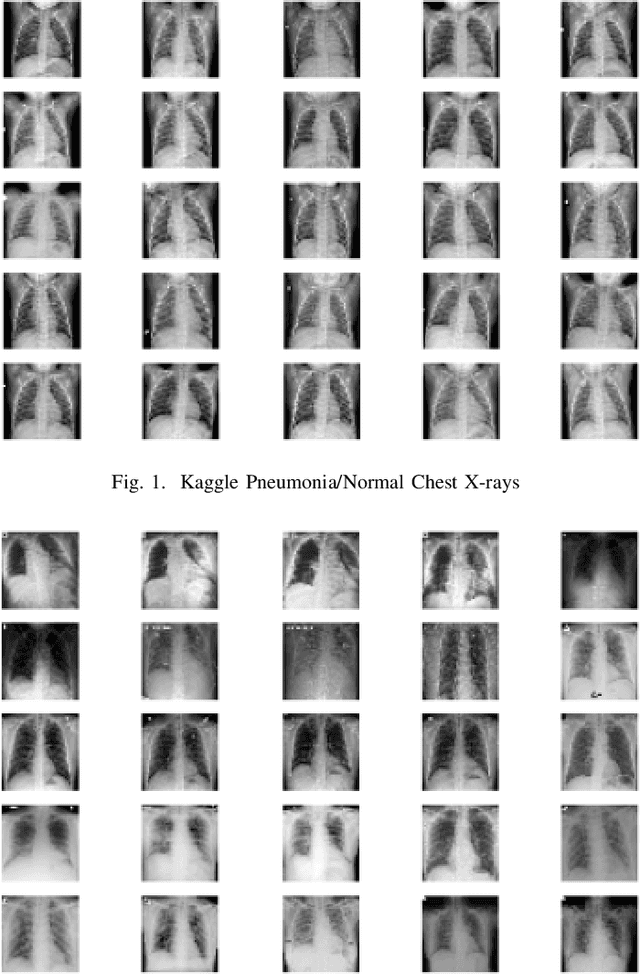
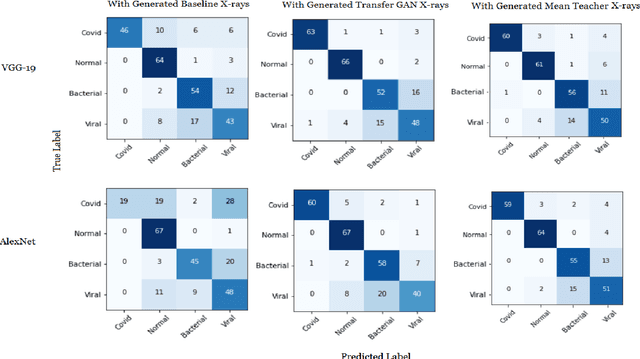
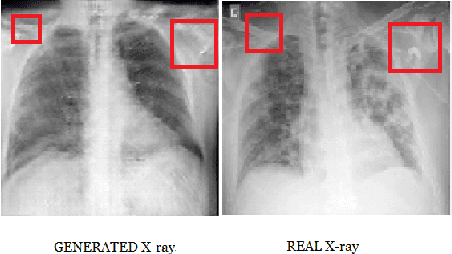
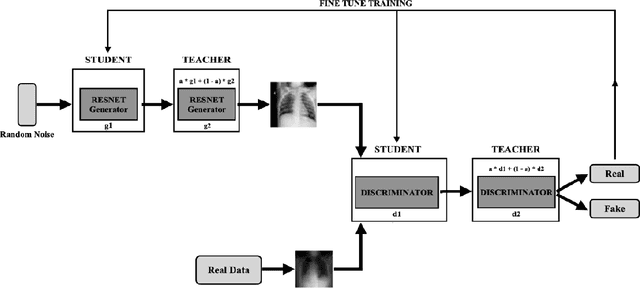
Abstract:COVID-19 is a novel infectious disease responsible for over 800K deaths worldwide as of August 2020. The need for rapid testing is a high priority and alternative testing strategies including X-ray image classification are a promising area of research. However, at present, public datasets for COVID19 x-ray images have low data volumes, making it challenging to develop accurate image classifiers. Several recent papers have made use of Generative Adversarial Networks (GANs) in order to increase the training data volumes. But realistic synthetic COVID19 X-rays remain challenging to generate. We present a novel Mean Teacher + Transfer GAN (MTT-GAN) that generates COVID19 chest X-ray images of high quality. In order to create a more accurate GAN, we employ transfer learning from the Kaggle Pneumonia X-Ray dataset, a highly relevant data source orders of magnitude larger than public COVID19 datasets. Furthermore, we employ the Mean Teacher algorithm as a constraint to improve stability of training. Our qualitative analysis shows that the MTT-GAN generates X-ray images that are greatly superior to a baseline GAN and visually comparable to real X-rays. Although board-certified radiologists can distinguish MTT-GAN fakes from real COVID19 X-rays. Quantitative analysis shows that MTT-GAN greatly improves the accuracy of both a binary COVID19 classifier as well as a multi-class Pneumonia classifier as compared to a baseline GAN. Our classification accuracy is favourable as compared to recently reported results in the literature for similar binary and multi-class COVID19 screening tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge