Miao Le
The Liver Tumor Segmentation Benchmark (LiTS)
Jan 13, 2019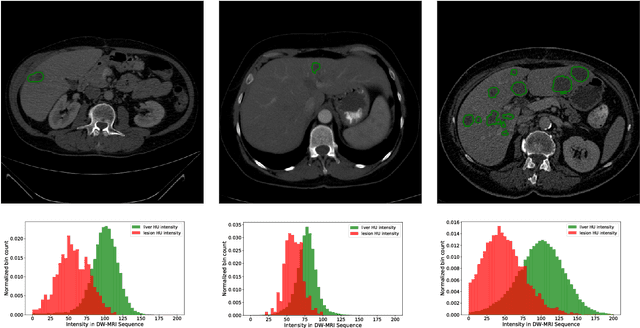
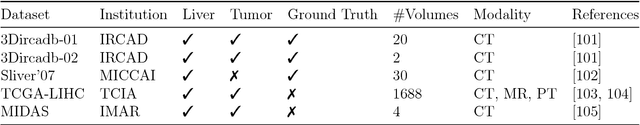
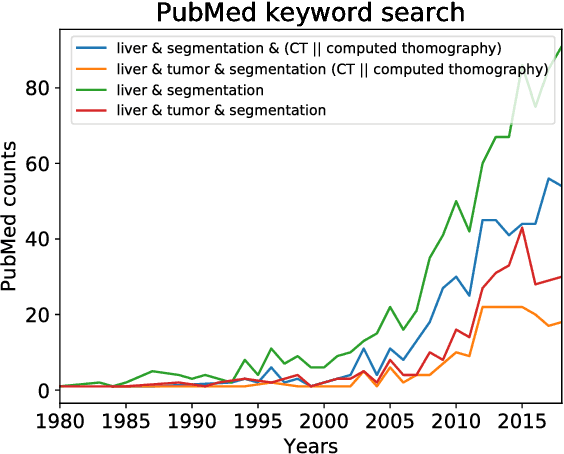
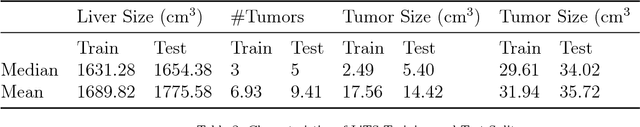
Abstract:In this work, we report the set-up and results of the Liver Tumor Segmentation Benchmark (LITS) organized in conjunction with the IEEE International Symposium on Biomedical Imaging (ISBI) 2016 and International Conference On Medical Image Computing Computer Assisted Intervention (MICCAI) 2017. Twenty four valid state-of-the-art liver and liver tumor segmentation algorithms were applied to a set of 131 computed tomography (CT) volumes with different types of tumor contrast levels (hyper-/hypo-intense), abnormalities in tissues (metastasectomie) size and varying amount of lesions. The submitted algorithms have been tested on 70 undisclosed volumes. The dataset is created in collaboration with seven hospitals and research institutions and manually reviewed by independent three radiologists. We found that not a single algorithm performed best for liver and tumors. The best liver segmentation algorithm achieved a Dice score of 0.96(MICCAI) whereas for tumor segmentation the best algorithm evaluated at 0.67(ISBI) and 0.70(MICCAI). The LITS image data and manual annotations continue to be publicly available through an online evaluation system as an ongoing benchmarking resource.
Global and Local Information Based Deep Network for Skin Lesion Segmentation
Mar 16, 2017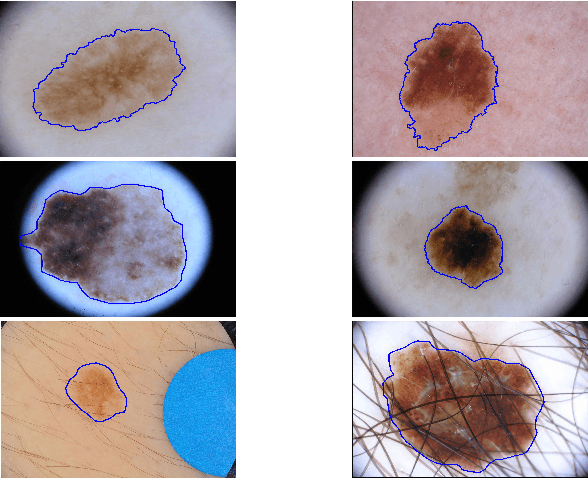
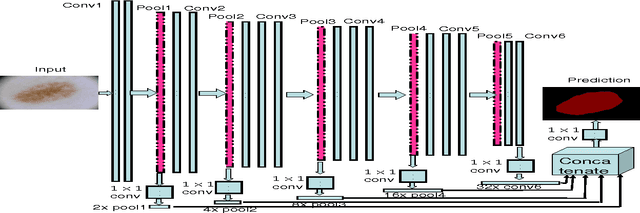
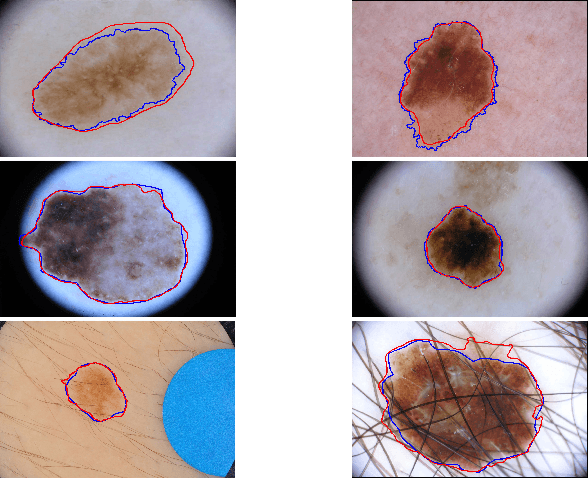
Abstract:With a large influx of dermoscopy images and a growing shortage of dermatologists, automatic dermoscopic image analysis plays an essential role in skin cancer diagnosis. In this paper, a new deep fully convolutional neural network (FCNN) is proposed to automatically segment melanoma out of skin images by end-to-end learning with only pixels and labels as inputs. Our proposed FCNN is capable of using both local and global information to segment melanoma by adopting skipping layers. The public benchmark database consisting of 150 validation images, 600 test images and 2000 training images in the melanoma detection challenge 2017 at International Symposium Biomedical Imaging 2017 is used to test the performance of our algorithm. All large size images (for example, $4000\times 6000$ pixels) are reduced to much smaller images with $384\times 384$ pixels (more than 10 times smaller). We got and submitted preliminary results to the challenge without any pre or post processing. The performance of our proposed method could be further improved by data augmentation and by avoiding image size reduction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge