Maxim Ziatdinov
Curiosity Driven Exploration to Optimize Structure-Property Learning in Microscopy
Apr 28, 2025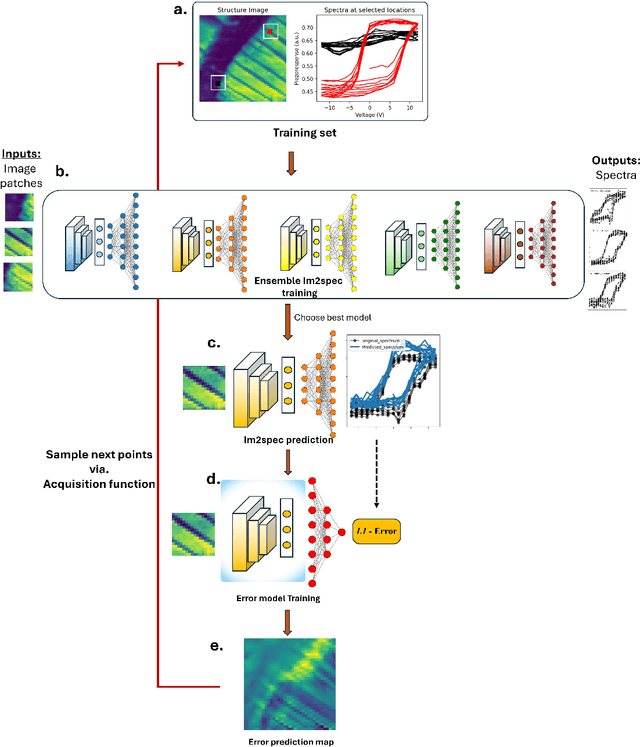
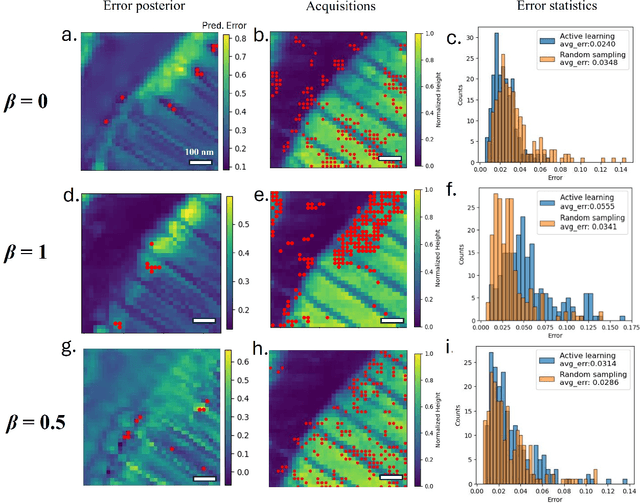
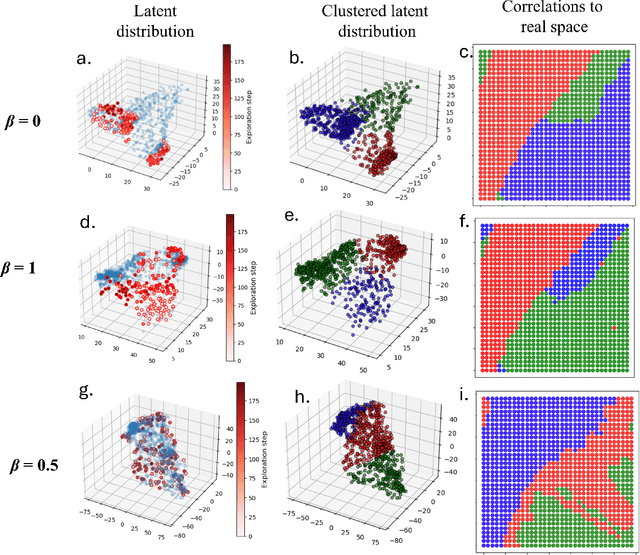
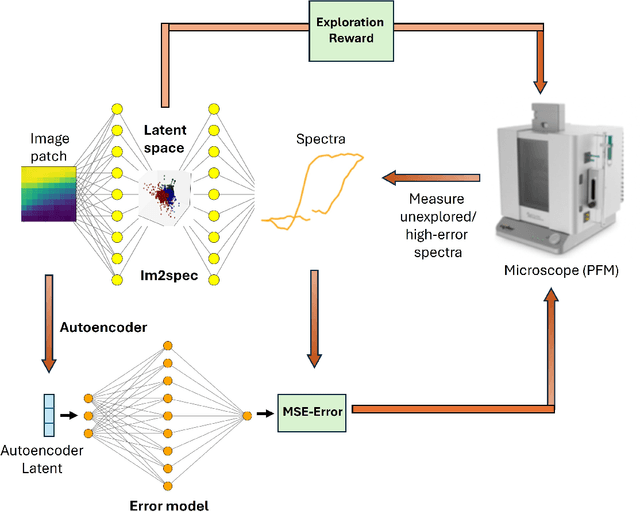
Abstract:Rapidly determining structure-property correlations in materials is an important challenge in better understanding fundamental mechanisms and greatly assists in materials design. In microscopy, imaging data provides a direct measurement of the local structure, while spectroscopic measurements provide relevant functional property information. Deep kernel active learning approaches have been utilized to rapidly map local structure to functional properties in microscopy experiments, but are computationally expensive for multi-dimensional and correlated output spaces. Here, we present an alternative lightweight curiosity algorithm which actively samples regions with unexplored structure-property relations, utilizing a deep-learning based surrogate model for error prediction. We show that the algorithm outperforms random sampling for predicting properties from structures, and provides a convenient tool for efficient mapping of structure-property relationships in materials science.
Automated Materials Discovery Platform Realized: Scanning Probe Microscopy of Combinatorial Libraries
Dec 24, 2024Abstract:Combinatorial libraries are a powerful approach for exploring the evolution of physical properties across binary and ternary cross-sections in multicomponent phase diagrams. Although the synthesis of these libraries has been developed since the 1960s and expedited with advanced laboratory automation, the broader application of combinatorial libraries relies on fast, reliable measurements of concentration-dependent structures and functionalities. Scanning Probe Microscopies (SPM), including piezoresponse force microscopy (PFM), offer significant potential for quantitative, functionally relevant combi-library readouts. Here we demonstrate the implementation of fully automated SPM to explore the evolution of ferroelectric properties in combinatorial libraries, focusing on Sm-doped BiFeO3 and ZnxMg1-xO systems. We also present and compare Gaussian Process-based Bayesian Optimization models for fully automated exploration, emphasizing local reproducibility (effective noise) as an essential factor in optimal experiment workflows. Automated SPM, when coupled with upstream synthesis controls, plays a pivotal role in bridging materials synthesis and characterization.
Predicting Battery Capacity Fade Using Probabilistic Machine Learning Models With and Without Pre-Trained Priors
Oct 08, 2024Abstract:Lithium-ion batteries are a key energy storage technology driving revolutions in mobile electronics, electric vehicles and renewable energy storage. Capacity retention is a vital performance measure that is frequently utilized to assess whether these batteries have approached their end-of-life. Machine learning (ML) offers a powerful tool for predicting capacity degradation based on past data, and, potentially, prior physical knowledge, but the degree to which an ML prediction can be trusted is of significant practical importance in situations where consequential decisions must be made based on battery state of health. This study explores the efficacy of fully Bayesian machine learning in forecasting battery health with the quantification of uncertainty in its predictions. Specifically, we implemented three probabilistic ML approaches and evaluated the accuracy of their predictions and uncertainty estimates: a standard Gaussian process (GP), a structured Gaussian process (sGP), and a fully Bayesian neural network (BNN). In typical applications of GP and sGP, their hyperparameters are learned from a single sample while, in contrast, BNNs are typically pre-trained on an existing dataset to learn the weight distributions before being used for inference. This difference in methodology gives the BNN an advantage in learning global trends in a dataset and makes BNNs a good choice when training data is available. However, we show that pre-training can also be leveraged for GP and sGP approaches to learn the prior distributions of the hyperparameters and that in the case of the pre-trained sGP, similar accuracy and improved uncertainty estimation compared to the BNN can be achieved. This approach offers a framework for a broad range of probabilistic machine learning scenarios where past data is available and can be used to learn priors for (hyper)parameters of probabilistic ML models.
Measurements with Noise: Bayesian Optimization for Co-optimizing Noise and Property Discovery in Automated Experiments
Oct 03, 2024Abstract:We have developed a Bayesian optimization (BO) workflow that integrates intra-step noise optimization into automated experimental cycles. Traditional BO approaches in automated experiments focus on optimizing experimental trajectories but often overlook the impact of measurement noise on data quality and cost. Our proposed framework simultaneously optimizes both the target property and the associated measurement noise by introducing time as an additional input parameter, thereby balancing the signal-to-noise ratio and experimental duration. Two approaches are explored: a reward-driven noise optimization and a double-optimization acquisition function, both enhancing the efficiency of automated workflows by considering noise and cost within the optimization process. We validate our method through simulations and real-world experiments using Piezoresponse Force Microscopy (PFM), demonstrating the successful optimization of measurement duration and property exploration. Our approach offers a scalable solution for optimizing multiple variables in automated experimental workflows, improving data quality, and reducing resource expenditure in materials science and beyond.
Active Learning with Fully Bayesian Neural Networks for Discontinuous and Nonstationary Data
May 17, 2024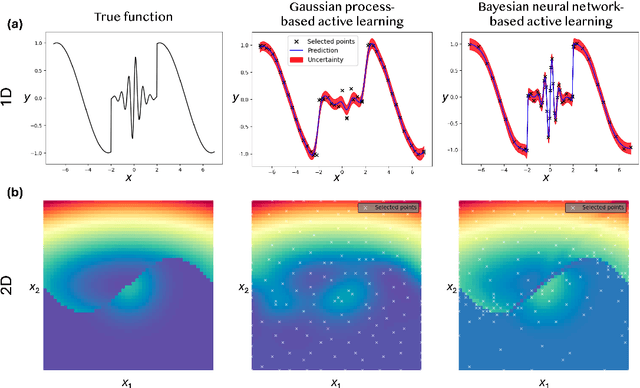
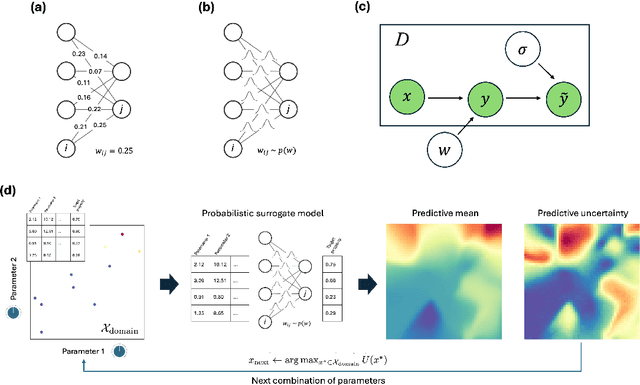
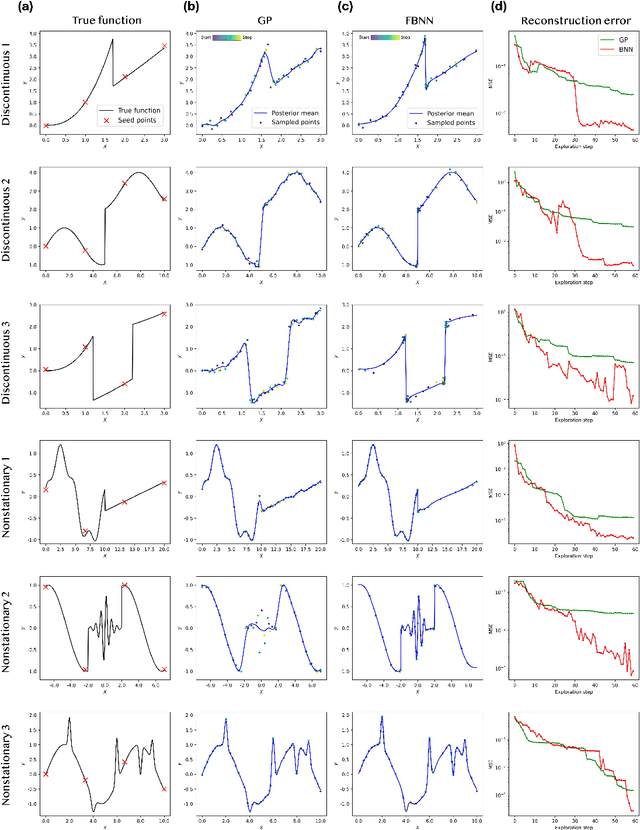
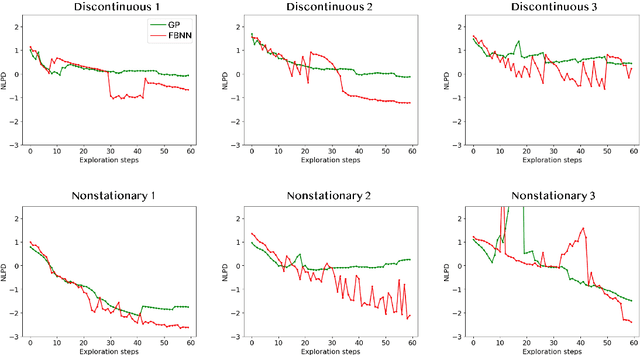
Abstract:Active learning optimizes the exploration of large parameter spaces by strategically selecting which experiments or simulations to conduct, thus reducing resource consumption and potentially accelerating scientific discovery. A key component of this approach is a probabilistic surrogate model, typically a Gaussian Process (GP), which approximates an unknown functional relationship between control parameters and a target property. However, conventional GPs often struggle when applied to systems with discontinuities and non-stationarities, prompting the exploration of alternative models. This limitation becomes particularly relevant in physical science problems, which are often characterized by abrupt transitions between different system states and rapid changes in physical property behavior. Fully Bayesian Neural Networks (FBNNs) serve as a promising substitute, treating all neural network weights probabilistically and leveraging advanced Markov Chain Monte Carlo techniques for direct sampling from the posterior distribution. This approach enables FBNNs to provide reliable predictive distributions, crucial for making informed decisions under uncertainty in the active learning setting. Although traditionally considered too computationally expensive for 'big data' applications, many physical sciences problems involve small amounts of data in relatively low-dimensional parameter spaces. Here, we assess the suitability and performance of FBNNs with the No-U-Turn Sampler for active learning tasks in the 'small data' regime, highlighting their potential to enhance predictive accuracy and reliability on test functions relevant to problems in physical sciences.
Towards accelerating physical discovery via non-interactive and interactive multi-fidelity Bayesian Optimization: Current challenges and future opportunities
Feb 20, 2024Abstract:Both computational and experimental material discovery bring forth the challenge of exploring multidimensional and often non-differentiable parameter spaces, such as phase diagrams of Hamiltonians with multiple interactions, composition spaces of combinatorial libraries, processing spaces, and molecular embedding spaces. Often these systems are expensive or time-consuming to evaluate a single instance, and hence classical approaches based on exhaustive grid or random search are too data intensive. This resulted in strong interest towards active learning methods such as Bayesian optimization (BO) where the adaptive exploration occurs based on human learning (discovery) objective. However, classical BO is based on a predefined optimization target, and policies balancing exploration and exploitation are purely data driven. In practical settings, the domain expert can pose prior knowledge on the system in form of partially known physics laws and often varies exploration policies during the experiment. Here, we explore interactive workflows building on multi-fidelity BO (MFBO), starting with classical (data-driven) MFBO, then structured (physics-driven) sMFBO, and extending it to allow human in the loop interactive iMFBO workflows for adaptive and domain expert aligned exploration. These approaches are demonstrated over highly non-smooth multi-fidelity simulation data generated from an Ising model, considering spin-spin interaction as parameter space, lattice sizes as fidelity spaces, and the objective as maximizing heat capacity. Detailed analysis and comparison show the impact of physics knowledge injection and on-the-fly human decisions for improved exploration, current challenges, and potential opportunities for algorithm development with combining data, physics and real time human decisions.
Human-in-the-loop: The future of Machine Learning in Automated Electron Microscopy
Oct 08, 2023Abstract:Machine learning methods are progressively gaining acceptance in the electron microscopy community for de-noising, semantic segmentation, and dimensionality reduction of data post-acquisition. The introduction of the APIs by major instrument manufacturers now allows the deployment of ML workflows in microscopes, not only for data analytics but also for real-time decision-making and feedback for microscope operation. However, the number of use cases for real-time ML remains remarkably small. Here, we discuss some considerations in designing ML-based active experiments and pose that the likely strategy for the next several years will be human-in-the-loop automated experiments (hAE). In this paradigm, the ML learning agent directly controls beam position and image and spectroscopy acquisition functions, and human operator monitors experiment progression in real- and feature space of the system and tunes the policies of the ML agent to steer the experiment towards specific objectives.
Towards Lightweight Data Integration using Multi-workflow Provenance and Data Observability
Aug 17, 2023



Abstract:Modern large-scale scientific discovery requires multidisciplinary collaboration across diverse computing facilities, including High Performance Computing (HPC) machines and the Edge-to-Cloud continuum. Integrated data analysis plays a crucial role in scientific discovery, especially in the current AI era, by enabling Responsible AI development, FAIR, Reproducibility, and User Steering. However, the heterogeneous nature of science poses challenges such as dealing with multiple supporting tools, cross-facility environments, and efficient HPC execution. Building on data observability, adapter system design, and provenance, we propose MIDA: an approach for lightweight runtime Multi-workflow Integrated Data Analysis. MIDA defines data observability strategies and adaptability methods for various parallel systems and machine learning tools. With observability, it intercepts the dataflows in the background without requiring instrumentation while integrating domain, provenance, and telemetry data at runtime into a unified database ready for user steering queries. We conduct experiments showing end-to-end multi-workflow analysis integrating data from Dask and MLFlow in a real distributed deep learning use case for materials science that runs on multiple environments with up to 276 GPUs in parallel. We show near-zero overhead running up to 100,000 tasks on 1,680 CPU cores on the Summit supercomputer.
* 10 pages, 5 figures, 2 Listings, 42 references, Paper accepted at IEEE eScience'23
Combining Variational Autoencoders and Physical Bias for Improved Microscopy Data Analysis
Feb 08, 2023Abstract:Electron and scanning probe microscopy produce vast amounts of data in the form of images or hyperspectral data, such as EELS or 4D STEM, that contain information on a wide range of structural, physical, and chemical properties of materials. To extract valuable insights from these data, it is crucial to identify physically separate regions in the data, such as phases, ferroic variants, and boundaries between them. In order to derive an easily interpretable feature analysis, combining with well-defined boundaries in a principled and unsupervised manner, here we present a physics augmented machine learning method which combines the capability of Variational Autoencoders to disentangle factors of variability within the data and the physics driven loss function that seeks to minimize the total length of the discontinuities in images corresponding to latent representations. Our method is applied to various materials, including NiO-LSMO, BiFeO3, and graphene. The results demonstrate the effectiveness of our approach in extracting meaningful information from large volumes of imaging data. The fully notebook containing implementation of the code and analysis workflow is available at https://github.com/arpanbiswas52/PaperNotebooks
Microscopy is All You Need
Oct 12, 2022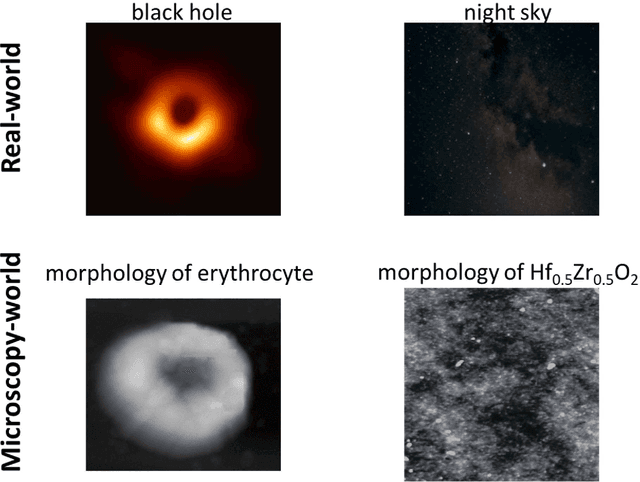
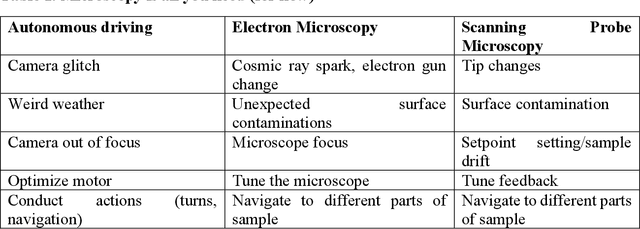
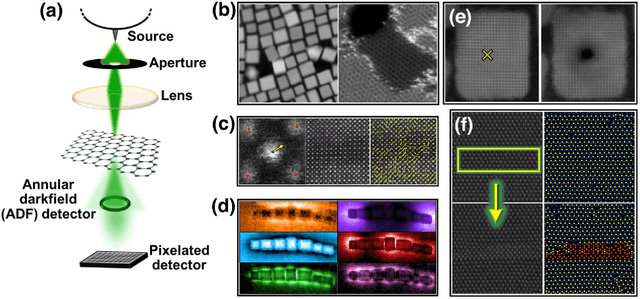
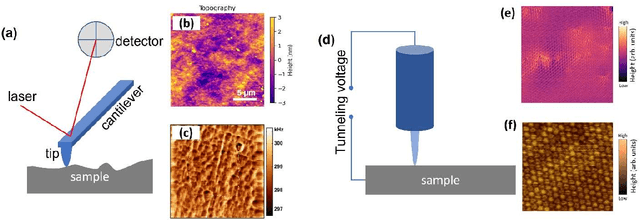
Abstract:We pose that microscopy offers an ideal real-world experimental environment for the development and deployment of active Bayesian and reinforcement learning methods. Indeed, the tremendous progress achieved by machine learning (ML) and artificial intelligence over the last decade has been largely achieved via the utilization of static data sets, from the paradigmatic MNIST to the bespoke corpora of text and image data used to train large models such as GPT3, DALLE and others. However, it is now recognized that continuous, minute improvements to state-of-the-art do not necessarily translate to advances in real-world applications. We argue that a promising pathway for the development of ML methods is via the route of domain-specific deployable algorithms in areas such as electron and scanning probe microscopy and chemical imaging. This will benefit both fundamental physical studies and serve as a test bed for more complex autonomous systems such as robotics and manufacturing. Favorable environment characteristics of scanning and electron microscopy include low risk, extensive availability of domain-specific priors and rewards, relatively small effects of exogeneous variables, and often the presence of both upstream first principles as well as downstream learnable physical models for both statics and dynamics. Recent developments in programmable interfaces, edge computing, and access to APIs facilitating microscope control, all render the deployment of ML codes on operational microscopes straightforward. We discuss these considerations and hope that these arguments will lead to creating a novel set of development targets for the ML community by accelerating both real-world ML applications and scientific progress.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge