Gerd Duscher
Towards Self-Optimizing Electron Microscope: Robust Tuning of Aberration Coefficients via Physics-Aware Multi-Objective Bayesian Optimization
Jan 26, 2026Abstract:Realizing high-throughput aberration-corrected Scanning Transmission Electron Microscopy (STEM) exploration of atomic structures requires rapid tuning of multipole probe correctors while compensating for the inevitable drift of the optical column. While automated alignment routines exist, conventional approaches rely on serial, gradient-free searches (e.g., Nelder-Mead) that are sample-inefficient and struggle to correct multiple interacting parameters simultaneously. Conversely, emerging deep learning methods offer speed but often lack the flexibility to adapt to varying sample conditions without extensive retraining. Here, we introduce a Multi-Objective Bayesian Optimization (MOBO) framework for rapid, data-efficient aberration correction. Importantly, this framework does not prescribe a single notion of image quality; instead, it enables user-defined, physically motivated reward formulations (e.g., symmetry-induced objectives) and uses Pareto fronts to expose the resulting trade-offs between competing experimental priorities. By using Gaussian Process regression to model the aberration landscape probabilistically, our workflow actively selects the most informative lens settings to evaluate next, rather than performing an exhaustive blind search. We demonstrate that this active learning loop is more robust than traditional optimization algorithms and effectively tunes focus, astigmatism, and higher-order aberrations. By balancing competing objectives, this approach enables "self-optimizing" microscopy by dynamically sustaining optimal performance during experiments.
Mic-hackathon 2024: Hackathon on Machine Learning for Electron and Scanning Probe Microscopy
Jun 10, 2025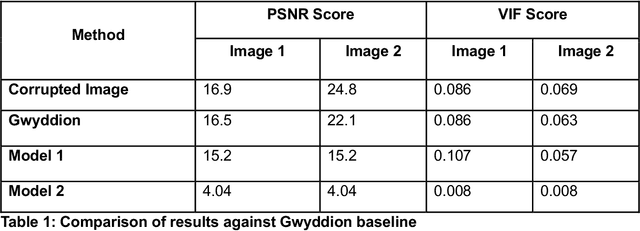
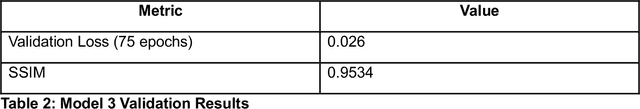
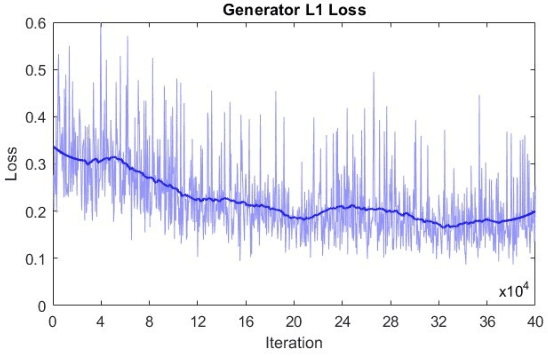
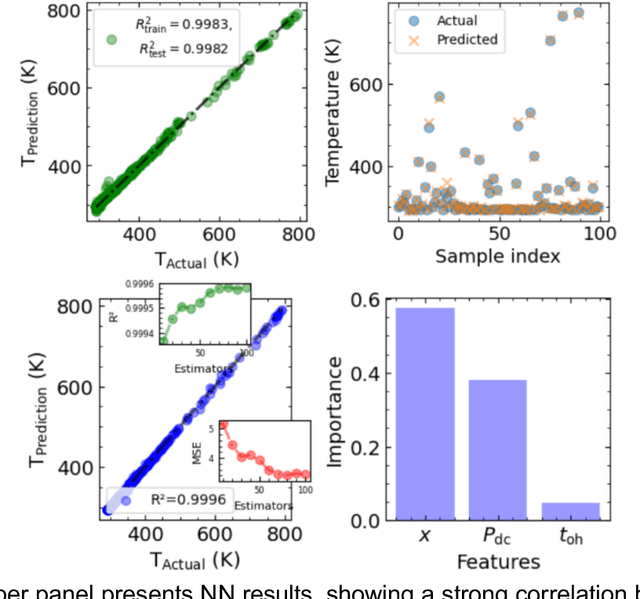
Abstract:Microscopy is a primary source of information on materials structure and functionality at nanometer and atomic scales. The data generated is often well-structured, enriched with metadata and sample histories, though not always consistent in detail or format. The adoption of Data Management Plans (DMPs) by major funding agencies promotes preservation and access. However, deriving insights remains difficult due to the lack of standardized code ecosystems, benchmarks, and integration strategies. As a result, data usage is inefficient and analysis time is extensive. In addition to post-acquisition analysis, new APIs from major microscope manufacturers enable real-time, ML-based analytics for automated decision-making and ML-agent-controlled microscope operation. Yet, a gap remains between the ML and microscopy communities, limiting the impact of these methods on physics, materials discovery, and optimization. Hackathons help bridge this divide by fostering collaboration between ML researchers and microscopy experts. They encourage the development of novel solutions that apply ML to microscopy, while preparing a future workforce for instrumentation, materials science, and applied ML. This hackathon produced benchmark datasets and digital twins of microscopes to support community growth and standardized workflows. All related code is available at GitHub: https://github.com/KalininGroup/Mic-hackathon-2024-codes-publication/tree/1.0.0.1
Human-in-the-loop: The future of Machine Learning in Automated Electron Microscopy
Oct 08, 2023Abstract:Machine learning methods are progressively gaining acceptance in the electron microscopy community for de-noising, semantic segmentation, and dimensionality reduction of data post-acquisition. The introduction of the APIs by major instrument manufacturers now allows the deployment of ML workflows in microscopes, not only for data analytics but also for real-time decision-making and feedback for microscope operation. However, the number of use cases for real-time ML remains remarkably small. Here, we discuss some considerations in designing ML-based active experiments and pose that the likely strategy for the next several years will be human-in-the-loop automated experiments (hAE). In this paradigm, the ML learning agent directly controls beam position and image and spectroscopy acquisition functions, and human operator monitors experiment progression in real- and feature space of the system and tunes the policies of the ML agent to steer the experiment towards specific objectives.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge