Kevin Roccapriore
Human-in-the-loop: The future of Machine Learning in Automated Electron Microscopy
Oct 08, 2023Abstract:Machine learning methods are progressively gaining acceptance in the electron microscopy community for de-noising, semantic segmentation, and dimensionality reduction of data post-acquisition. The introduction of the APIs by major instrument manufacturers now allows the deployment of ML workflows in microscopes, not only for data analytics but also for real-time decision-making and feedback for microscope operation. However, the number of use cases for real-time ML remains remarkably small. Here, we discuss some considerations in designing ML-based active experiments and pose that the likely strategy for the next several years will be human-in-the-loop automated experiments (hAE). In this paradigm, the ML learning agent directly controls beam position and image and spectroscopy acquisition functions, and human operator monitors experiment progression in real- and feature space of the system and tunes the policies of the ML agent to steer the experiment towards specific objectives.
Microscopy is All You Need
Oct 12, 2022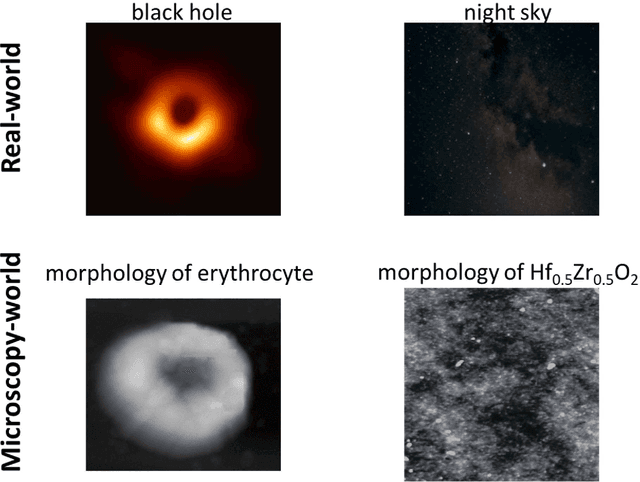
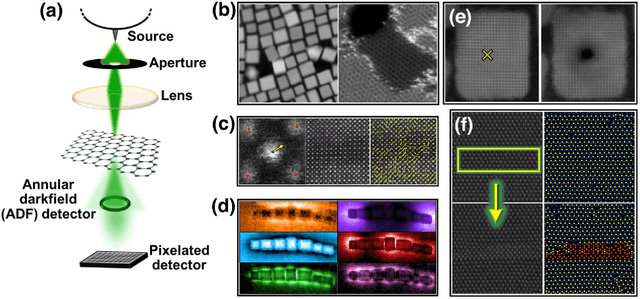
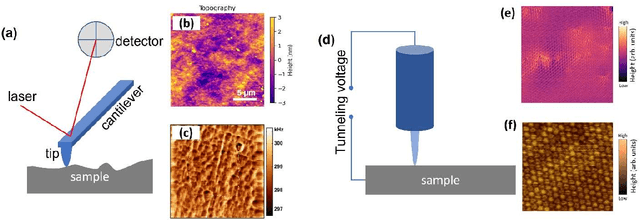
Abstract:We pose that microscopy offers an ideal real-world experimental environment for the development and deployment of active Bayesian and reinforcement learning methods. Indeed, the tremendous progress achieved by machine learning (ML) and artificial intelligence over the last decade has been largely achieved via the utilization of static data sets, from the paradigmatic MNIST to the bespoke corpora of text and image data used to train large models such as GPT3, DALLE and others. However, it is now recognized that continuous, minute improvements to state-of-the-art do not necessarily translate to advances in real-world applications. We argue that a promising pathway for the development of ML methods is via the route of domain-specific deployable algorithms in areas such as electron and scanning probe microscopy and chemical imaging. This will benefit both fundamental physical studies and serve as a test bed for more complex autonomous systems such as robotics and manufacturing. Favorable environment characteristics of scanning and electron microscopy include low risk, extensive availability of domain-specific priors and rewards, relatively small effects of exogeneous variables, and often the presence of both upstream first principles as well as downstream learnable physical models for both statics and dynamics. Recent developments in programmable interfaces, edge computing, and access to APIs facilitating microscope control, all render the deployment of ML codes on operational microscopes straightforward. We discuss these considerations and hope that these arguments will lead to creating a novel set of development targets for the ML community by accelerating both real-world ML applications and scientific progress.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge