Matthias Perkonigg
Improving Vessel Segmentation with Multi-Task Learning and Auxiliary Data Available Only During Model Training
Sep 04, 2025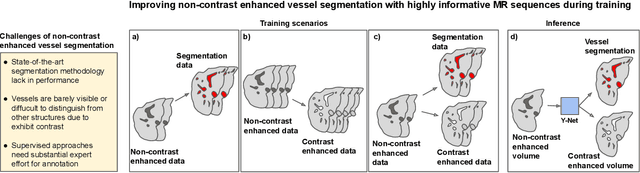

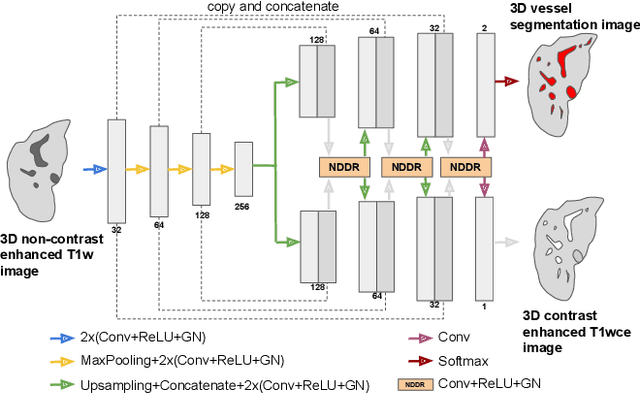

Abstract:Liver vessel segmentation in magnetic resonance imaging data is important for the computational analysis of vascular remodelling, associated with a wide spectrum of diffuse liver diseases. Existing approaches rely on contrast enhanced imaging data, but the necessary dedicated imaging sequences are not uniformly acquired. Images without contrast enhancement are acquired more frequently, but vessel segmentation is challenging, and requires large-scale annotated data. We propose a multi-task learning framework to segment vessels in liver MRI without contrast. It exploits auxiliary contrast enhanced MRI data available only during training to reduce the need for annotated training examples. Our approach draws on paired native and contrast enhanced data with and without vessel annotations for model training. Results show that auxiliary data improves the accuracy of vessel segmentation, even if they are not available during inference. The advantage is most pronounced if only few annotations are available for training, since the feature representation benefits from the shared task structure. A validation of this approach to augment a model for brain tumor segmentation confirms its benefits across different domains. An auxiliary informative imaging modality can augment expert annotations even if it is only available during training.
Identifying Signatures of Image Phenotypes to Track Treatment Response in Liver Disease
Jul 16, 2025



Abstract:Quantifiable image patterns associated with disease progression and treatment response are critical tools for guiding individual treatment, and for developing novel therapies. Here, we show that unsupervised machine learning can identify a pattern vocabulary of liver tissue in magnetic resonance images that quantifies treatment response in diffuse liver disease. Deep clustering networks simultaneously encode and cluster patches of medical images into a low-dimensional latent space to establish a tissue vocabulary. The resulting tissue types capture differential tissue change and its location in the liver associated with treatment response. We demonstrate the utility of the vocabulary on a randomized controlled trial cohort of non-alcoholic steatohepatitis patients. First, we use the vocabulary to compare longitudinal liver change in a placebo and a treatment cohort. Results show that the method identifies specific liver tissue change pathways associated with treatment, and enables a better separation between treatment groups than established non-imaging measures. Moreover, we show that the vocabulary can predict biopsy derived features from non-invasive imaging data. We validate the method on a separate replication cohort to demonstrate the applicability of the proposed method.
Continual Active Learning Using Pseudo-Domains for Limited Labelling Resources and Changing Acquisition Characteristics
Nov 25, 2021



Abstract:Machine learning in medical imaging during clinical routine is impaired by changes in scanner protocols, hardware, or policies resulting in a heterogeneous set of acquisition settings. When training a deep learning model on an initial static training set, model performance and reliability suffer from changes of acquisition characteristics as data and targets may become inconsistent. Continual learning can help to adapt models to the changing environment by training on a continuous data stream. However, continual manual expert labelling of medical imaging requires substantial effort. Thus, ways to use labelling resources efficiently on a well chosen sub-set of new examples is necessary to render this strategy feasible. Here, we propose a method for continual active learning operating on a stream of medical images in a multi-scanner setting. The approach automatically recognizes shifts in image acquisition characteristics - new domains -, selects optimal examples for labelling and adapts training accordingly. Labelling is subject to a limited budget, resembling typical real world scenarios. To demonstrate generalizability, we evaluate the effectiveness of our method on three tasks: cardiac segmentation, lung nodule detection and brain age estimation. Results show that the proposed approach outperforms other active learning methods, while effectively counteracting catastrophic forgetting.
Pseudo-domains in imaging data improve prediction of future disease status in multi-center studies
Nov 15, 2021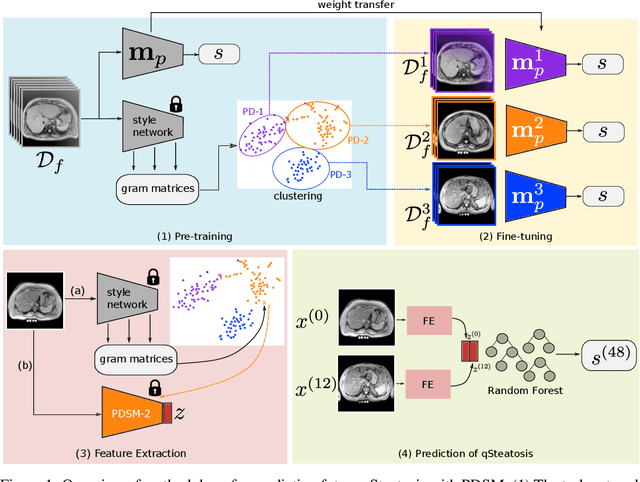
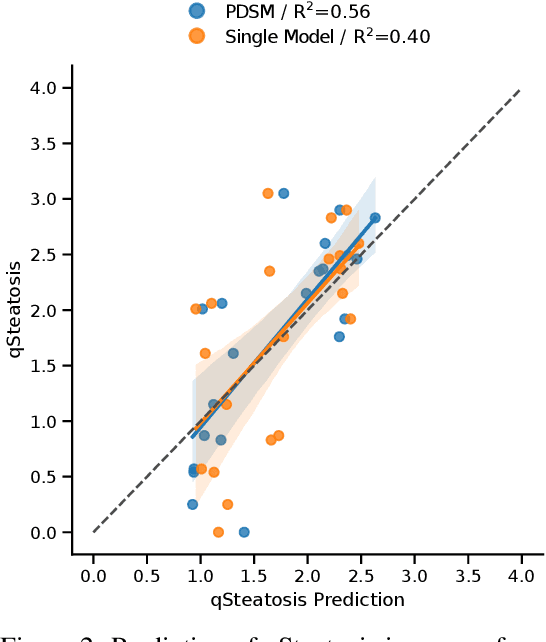
Abstract:In multi-center randomized clinical trials imaging data can be diverse due to acquisition technology or scanning protocols. Models predicting future outcome of patients are impaired by this data heterogeneity. Here, we propose a prediction method that can cope with a high number of different scanning sites and a low number of samples per site. We cluster sites into pseudo-domains based on visual appearance of scans, and train pseudo-domain specific models. Results show that they improve the prediction accuracy for steatosis after 48 weeks from imaging data acquired at an initial visit and 12-weeks follow-up in liver disease
Continual Active Learning for Efficient Adaptation of Machine Learning Models to Changing Image Acquisition
Jun 07, 2021



Abstract:Imaging in clinical routine is subject to changing scanner protocols, hardware, or policies in a typically heterogeneous set of acquisition hardware. Accuracy and reliability of deep learning models suffer from those changes as data and targets become inconsistent with their initial static training set. Continual learning can adapt to a continuous data stream of a changing imaging environment. Here, we propose a method for continual active learning on a data stream of medical images. It recognizes shifts or additions of new imaging sources - domains -, adapts training accordingly, and selects optimal examples for labelling. Model training has to cope with a limited labelling budget, resembling typical real world scenarios. We demonstrate our method on T1-weighted magnetic resonance images from three different scanners with the task of brain age estimation. Results demonstrate that the proposed method outperforms naive active learning while requiring less manual labelling.
Dynamic memory to alleviate catastrophic forgetting in continuous learning settings
Jul 07, 2020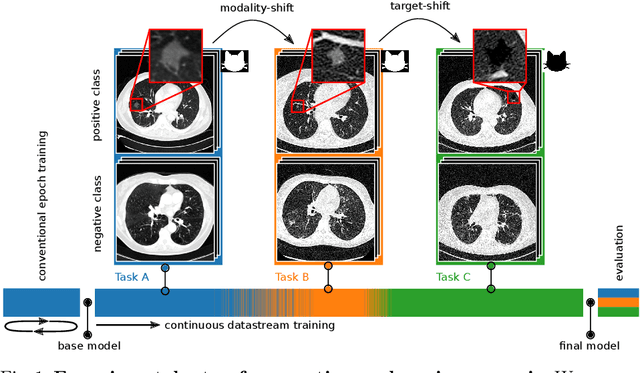

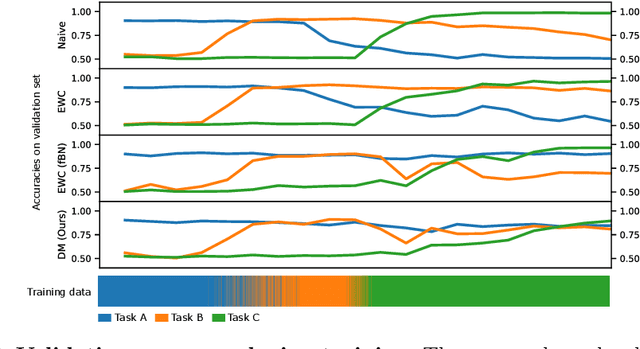
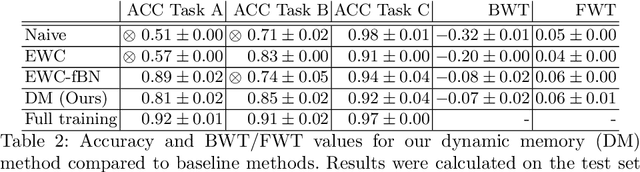
Abstract:In medical imaging, technical progress or changes in diagnostic procedures lead to a continuous change in image appearance. Scanner manufacturer, reconstruction kernel, dose, other protocol specific settings or administering of contrast agents are examples that influence image content independent of the scanned biology. Such domain and task shifts limit the applicability of machine learning algorithms in the clinical routine by rendering models obsolete over time. Here, we address the problem of data shifts in a continuous learning scenario by adapting a model to unseen variations in the source domain while counteracting catastrophic forgetting effects. Our method uses a dynamic memory to facilitate rehearsal of a diverse training data subset to mitigate forgetting. We evaluated our approach on routine clinical CT data obtained with two different scanner protocols and synthetic classification tasks. Experiments show that dynamic memory counters catastrophic forgetting in a setting with multiple data shifts without the necessity for explicit knowledge about when these shifts occur.
Unsupervised deep clustering for predictive texture pattern discovery in medical images
Jan 31, 2020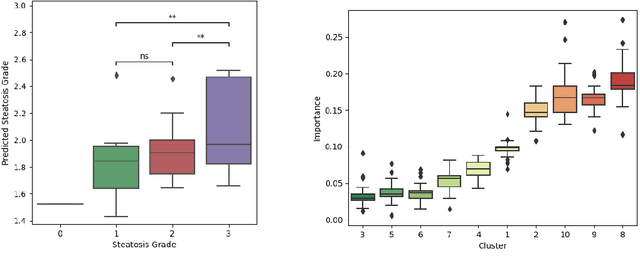
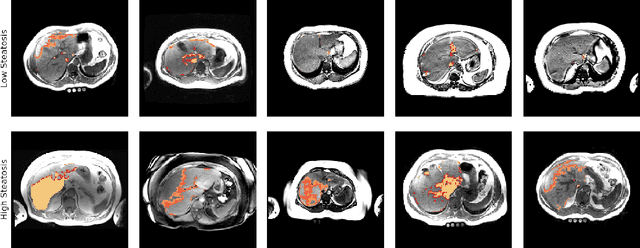
Abstract:Predictive marker patterns in imaging data are a means to quantify disease and progression, but their identification is challenging, if the underlying biology is poorly understood. Here, we present a method to identify predictive texture patterns in medical images in an unsupervised way. Based on deep clustering networks, we simultaneously encode and cluster medical image patches in a low-dimensional latent space. The resulting clusters serve as features for disease staging, linking them to the underlying disease. We evaluate the method on 70 T1-weighted magnetic resonance images of patients with different stages of liver steatosis. The deep clustering approach is able to find predictive clusters with a stable ranking, differentiating between low and high steatosis with an F1-Score of 0.78.
CHAOS Challenge -- Combined (CT-MR) Healthy Abdominal Organ Segmentation
Jan 17, 2020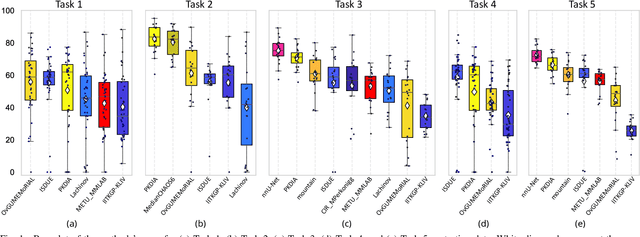
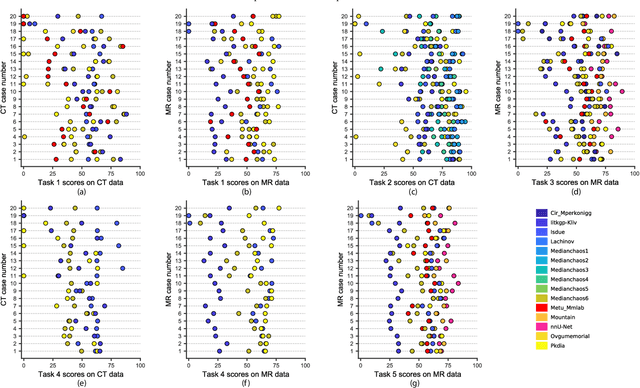
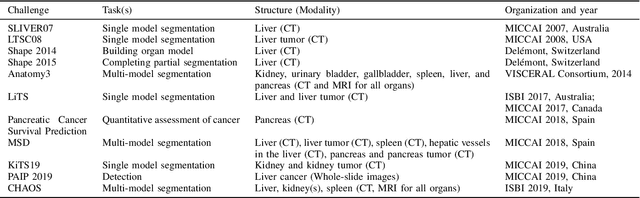
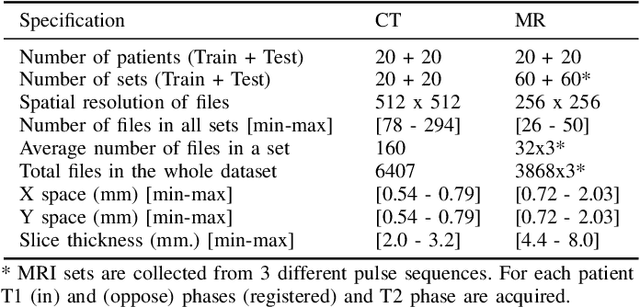
Abstract:Segmentation of abdominal organs has been a comprehensive, yet unresolved, research field for many years. In the last decade, intensive developments in deep learning (DL) have introduced new state-of-the-art segmentation systems. Despite outperforming the overall accuracy of existing systems, the effects of DL model properties and parameters on the performance is hard to interpret. This makes comparative analysis a necessary tool to achieve explainable studies and systems. Moreover, the performance of DL for emerging learning approaches such as cross-modality and multi-modal tasks have been rarely discussed. In order to expand the knowledge in these topics, CHAOS -- Combined (CT-MR) Healthy Abdominal Organ Segmentation challenge has been organized in the IEEE International Symposium on Biomedical Imaging (ISBI), 2019, in Venice, Italy. Despite a large number of the previous abdomen related challenges, the majority of which are focused on tumor/lesion detection and/or classification with a single modality, CHAOS provides both abdominal CT and MR data from healthy subjects. Five different and complementary tasks have been designed to analyze the capabilities of the current approaches from multiple perspectives. The results are investigated thoroughly, compared with manual annotations and interactive methods. The outcomes are reported in detail to reflect the latest advancements in the field. CHAOS challenge and data will be available online to provide a continuous benchmark resource for segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge