Ahmed Ba-Ssalamah
MRI-derived quantification of hepatic vessel-to-volume ratios in chronic liver disease using a deep learning approach
Oct 09, 2025Abstract:Background: We aimed to quantify hepatic vessel volumes across chronic liver disease stages and healthy controls using deep learning-based magnetic resonance imaging (MRI) analysis, and assess correlations with biomarkers for liver (dys)function and fibrosis/portal hypertension. Methods: We assessed retrospectively healthy controls, non-advanced and advanced chronic liver disease (ACLD) patients using a 3D U-Net model for hepatic vessel segmentation on portal venous phase gadoxetic acid-enhanced 3-T MRI. Total (TVVR), hepatic (HVVR), and intrahepatic portal vein-to-volume ratios (PVVR) were compared between groups and correlated with: albumin-bilirubin (ALBI) and model for end-stage liver disease-sodium (MELD-Na) score, and fibrosis/portal hypertension (Fibrosis-4 [FIB-4] score, liver stiffness measurement [LSM], hepatic venous pressure gradient [HVPG], platelet count [PLT], and spleen volume). Results: We included 197 subjects, aged 54.9 $\pm$ 13.8 years (mean $\pm$ standard deviation), 111 males (56.3\%): 35 healthy controls, 44 non-ACLD, and 118 ACLD patients. TVVR and HVVR were highest in controls (3.9; 2.1), intermediate in non-ACLD (2.8; 1.7), and lowest in ACLD patients (2.3; 1.0) ($p \leq 0.001$). PVVR was reduced in both non-ACLD and ACLD patients (both 1.2) compared to controls (1.7) ($p \leq 0.001$), but showed no difference between CLD groups ($p = 0.999$). HVVR significantly correlated indirectly with FIB-4, ALBI, MELD-Na, LSM, and spleen volume ($\rho$ ranging from -0.27 to -0.40), and directly with PLT ($\rho = 0.36$). TVVR and PVVR showed similar but weaker correlations. Conclusions: Deep learning-based hepatic vessel volumetry demonstrated differences between healthy liver and chronic liver disease stages and shows correlations with established markers of disease severity.
Improving Vessel Segmentation with Multi-Task Learning and Auxiliary Data Available Only During Model Training
Sep 04, 2025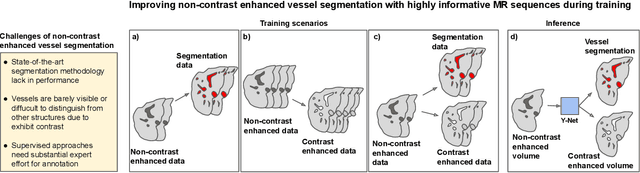

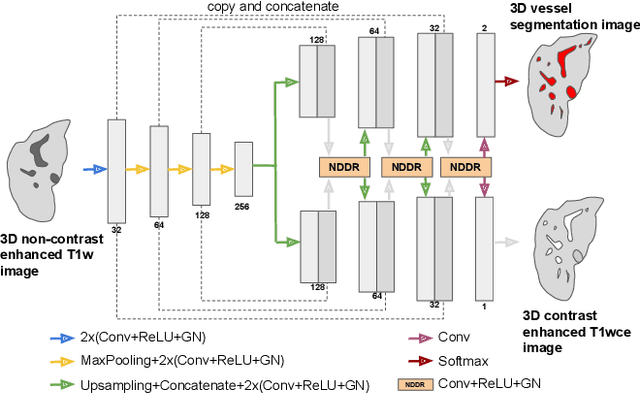

Abstract:Liver vessel segmentation in magnetic resonance imaging data is important for the computational analysis of vascular remodelling, associated with a wide spectrum of diffuse liver diseases. Existing approaches rely on contrast enhanced imaging data, but the necessary dedicated imaging sequences are not uniformly acquired. Images without contrast enhancement are acquired more frequently, but vessel segmentation is challenging, and requires large-scale annotated data. We propose a multi-task learning framework to segment vessels in liver MRI without contrast. It exploits auxiliary contrast enhanced MRI data available only during training to reduce the need for annotated training examples. Our approach draws on paired native and contrast enhanced data with and without vessel annotations for model training. Results show that auxiliary data improves the accuracy of vessel segmentation, even if they are not available during inference. The advantage is most pronounced if only few annotations are available for training, since the feature representation benefits from the shared task structure. A validation of this approach to augment a model for brain tumor segmentation confirms its benefits across different domains. An auxiliary informative imaging modality can augment expert annotations even if it is only available during training.
Identifying Signatures of Image Phenotypes to Track Treatment Response in Liver Disease
Jul 16, 2025



Abstract:Quantifiable image patterns associated with disease progression and treatment response are critical tools for guiding individual treatment, and for developing novel therapies. Here, we show that unsupervised machine learning can identify a pattern vocabulary of liver tissue in magnetic resonance images that quantifies treatment response in diffuse liver disease. Deep clustering networks simultaneously encode and cluster patches of medical images into a low-dimensional latent space to establish a tissue vocabulary. The resulting tissue types capture differential tissue change and its location in the liver associated with treatment response. We demonstrate the utility of the vocabulary on a randomized controlled trial cohort of non-alcoholic steatohepatitis patients. First, we use the vocabulary to compare longitudinal liver change in a placebo and a treatment cohort. Results show that the method identifies specific liver tissue change pathways associated with treatment, and enables a better separation between treatment groups than established non-imaging measures. Moreover, we show that the vocabulary can predict biopsy derived features from non-invasive imaging data. We validate the method on a separate replication cohort to demonstrate the applicability of the proposed method.
Pseudo-domains in imaging data improve prediction of future disease status in multi-center studies
Nov 15, 2021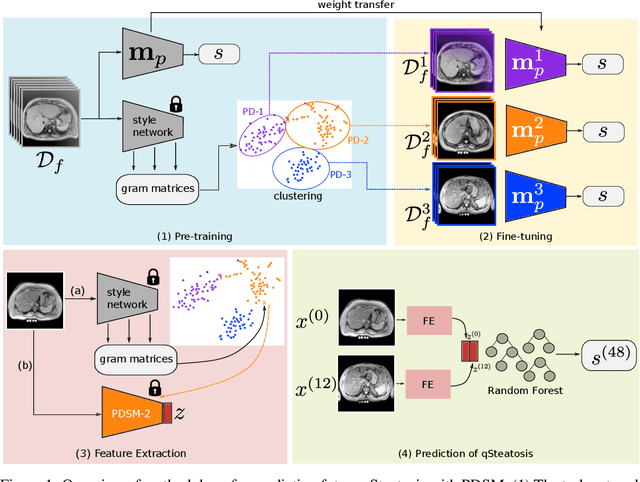
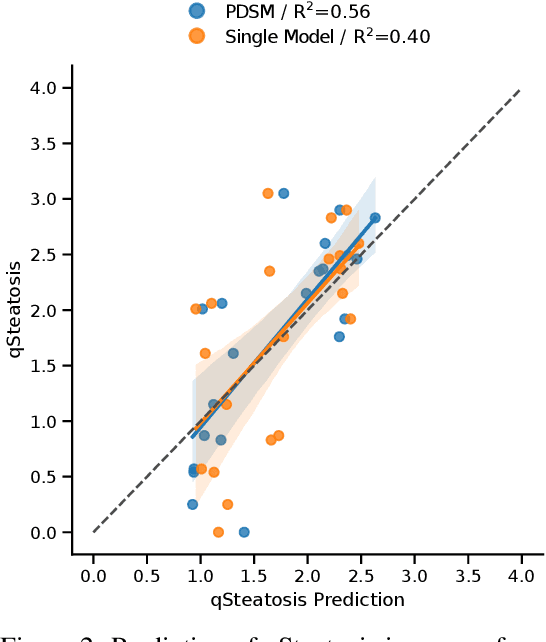
Abstract:In multi-center randomized clinical trials imaging data can be diverse due to acquisition technology or scanning protocols. Models predicting future outcome of patients are impaired by this data heterogeneity. Here, we propose a prediction method that can cope with a high number of different scanning sites and a low number of samples per site. We cluster sites into pseudo-domains based on visual appearance of scans, and train pseudo-domain specific models. Results show that they improve the prediction accuracy for steatosis after 48 weeks from imaging data acquired at an initial visit and 12-weeks follow-up in liver disease
Unsupervised deep clustering for predictive texture pattern discovery in medical images
Jan 31, 2020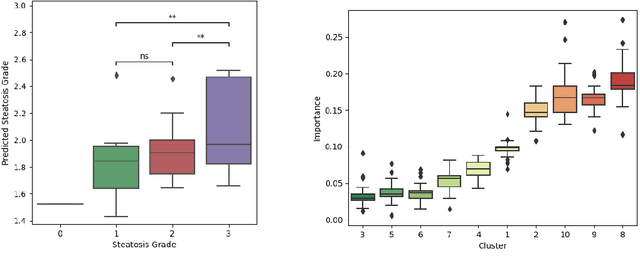
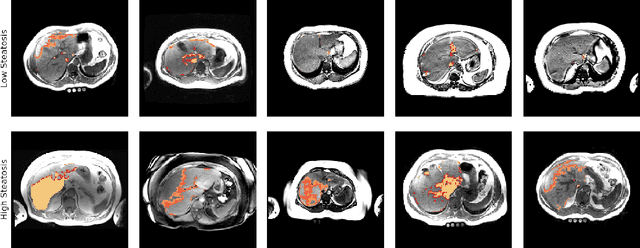
Abstract:Predictive marker patterns in imaging data are a means to quantify disease and progression, but their identification is challenging, if the underlying biology is poorly understood. Here, we present a method to identify predictive texture patterns in medical images in an unsupervised way. Based on deep clustering networks, we simultaneously encode and cluster medical image patches in a low-dimensional latent space. The resulting clusters serve as features for disease staging, linking them to the underlying disease. We evaluate the method on 70 T1-weighted magnetic resonance images of patients with different stages of liver steatosis. The deep clustering approach is able to find predictive clusters with a stable ranking, differentiating between low and high steatosis with an F1-Score of 0.78.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge