Johannes Hofmanninger
Continual Active Learning Using Pseudo-Domains for Limited Labelling Resources and Changing Acquisition Characteristics
Nov 25, 2021



Abstract:Machine learning in medical imaging during clinical routine is impaired by changes in scanner protocols, hardware, or policies resulting in a heterogeneous set of acquisition settings. When training a deep learning model on an initial static training set, model performance and reliability suffer from changes of acquisition characteristics as data and targets may become inconsistent. Continual learning can help to adapt models to the changing environment by training on a continuous data stream. However, continual manual expert labelling of medical imaging requires substantial effort. Thus, ways to use labelling resources efficiently on a well chosen sub-set of new examples is necessary to render this strategy feasible. Here, we propose a method for continual active learning operating on a stream of medical images in a multi-scanner setting. The approach automatically recognizes shifts in image acquisition characteristics - new domains -, selects optimal examples for labelling and adapts training accordingly. Labelling is subject to a limited budget, resembling typical real world scenarios. To demonstrate generalizability, we evaluate the effectiveness of our method on three tasks: cardiac segmentation, lung nodule detection and brain age estimation. Results show that the proposed approach outperforms other active learning methods, while effectively counteracting catastrophic forgetting.
Continual Active Learning for Efficient Adaptation of Machine Learning Models to Changing Image Acquisition
Jun 07, 2021



Abstract:Imaging in clinical routine is subject to changing scanner protocols, hardware, or policies in a typically heterogeneous set of acquisition hardware. Accuracy and reliability of deep learning models suffer from those changes as data and targets become inconsistent with their initial static training set. Continual learning can adapt to a continuous data stream of a changing imaging environment. Here, we propose a method for continual active learning on a data stream of medical images. It recognizes shifts or additions of new imaging sources - domains -, adapts training accordingly, and selects optimal examples for labelling. Model training has to cope with a limited labelling budget, resembling typical real world scenarios. We demonstrate our method on T1-weighted magnetic resonance images from three different scanners with the task of brain age estimation. Results demonstrate that the proposed method outperforms naive active learning while requiring less manual labelling.
Dynamic memory to alleviate catastrophic forgetting in continuous learning settings
Jul 07, 2020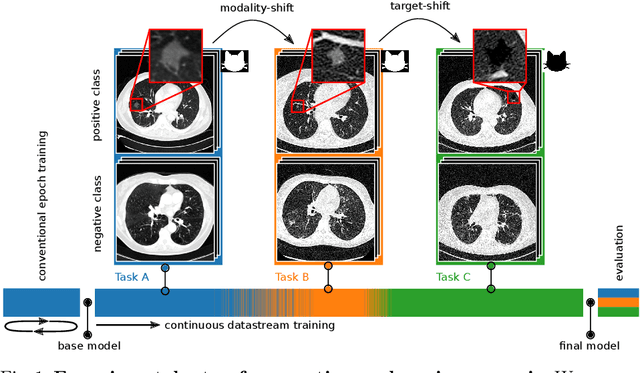

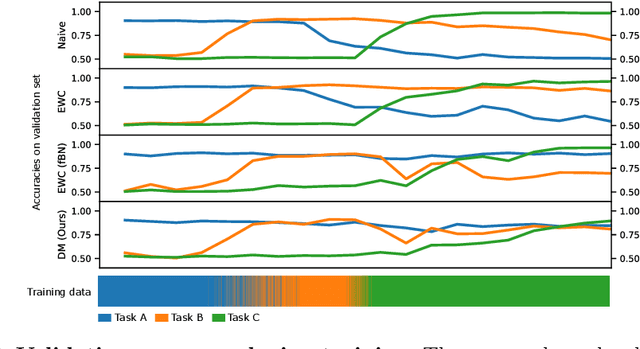
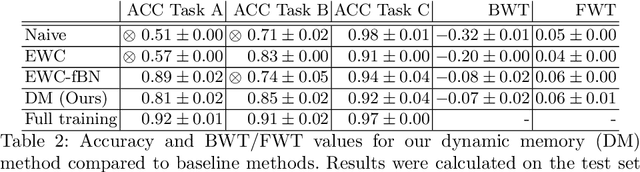
Abstract:In medical imaging, technical progress or changes in diagnostic procedures lead to a continuous change in image appearance. Scanner manufacturer, reconstruction kernel, dose, other protocol specific settings or administering of contrast agents are examples that influence image content independent of the scanned biology. Such domain and task shifts limit the applicability of machine learning algorithms in the clinical routine by rendering models obsolete over time. Here, we address the problem of data shifts in a continuous learning scenario by adapting a model to unseen variations in the source domain while counteracting catastrophic forgetting effects. Our method uses a dynamic memory to facilitate rehearsal of a diverse training data subset to mitigate forgetting. We evaluated our approach on routine clinical CT data obtained with two different scanner protocols and synthetic classification tasks. Experiments show that dynamic memory counters catastrophic forgetting in a setting with multiple data shifts without the necessity for explicit knowledge about when these shifts occur.
Separation of target anatomical structure and occlusions in chest radiographs
Feb 03, 2020
Abstract:Chest radiographs are commonly performed low-cost exams for screening and diagnosis. However, radiographs are 2D representations of 3D structures causing considerable clutter impeding visual inspection and automated image analysis. Here, we propose a Fully Convolutional Network to suppress, for a specific task, undesired visual structure from radiographs while retaining the relevant image information such as lung-parenchyma. The proposed algorithm creates reconstructed radiographs and ground-truth data from high resolution CT-scans. Results show that removing visual variation that is irrelevant for a classification task improves the performance of a classifier when only limited training data are available. This is particularly relevant because a low number of ground-truth cases is common in medical imaging.
Automatic lung segmentation in routine imaging is a data diversity problem, not a methodology problem
Jan 31, 2020



Abstract:Automated segmentation of anatomical structures is a crucial step in many medical image analysis tasks. For lung segmentation, a variety of approaches exist, involving sophisticated pipelines trained and validated on a range of different data sets. However, during translation to clinical routine the applicability of these approaches across diseases remains limited. Here, we show that the accuracy and reliability of lung segmentation algorithms on demanding cases primarily does not depend on methodology, but on the diversity of training data. We compare 4 generic deep learning approaches and 2 published lung segmentation algorithms on routine imaging data with more than 6 different disease patterns and 3 published data sets. We show that a basic approach - U-net - performs either better, or competitively with other approaches on both routine data and published data sets, and outperforms published approaches once trained on a diverse data set covering multiple diseases. Training data composition consistently has a bigger impact than algorithm choice on accuracy across test data sets. We carefully analyse the impact of data diversity, and the specifications of annotations on both training and validation sets to provide a reference for algorithms, training data, and annotation. Results on a seemingly well understood task of lung segmentation suggest the critical importance of training data diversity compared to model choice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge