N. Sinem Gezer
Cross-Domain Segmentation with Adversarial Loss and Covariate Shift for Biomedical Imaging
Jun 08, 2020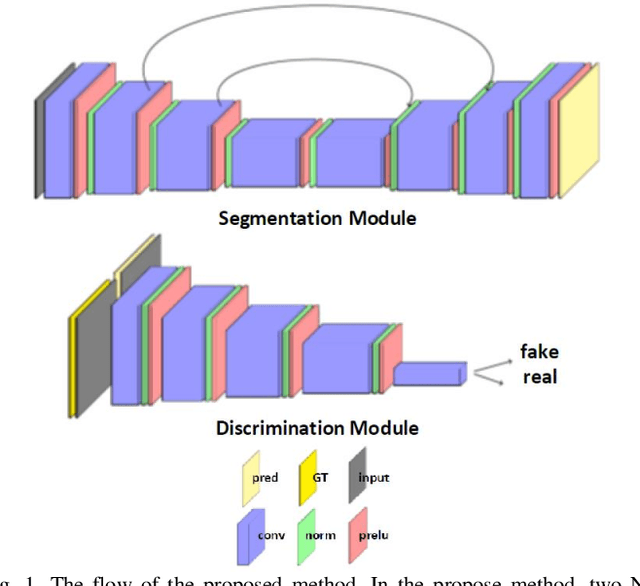
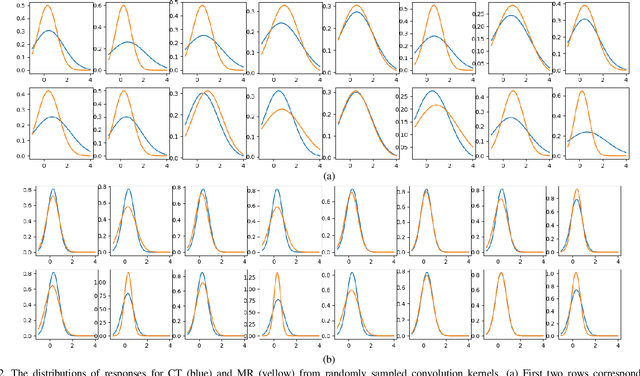
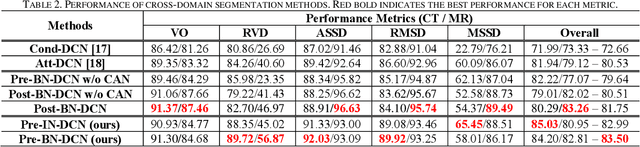
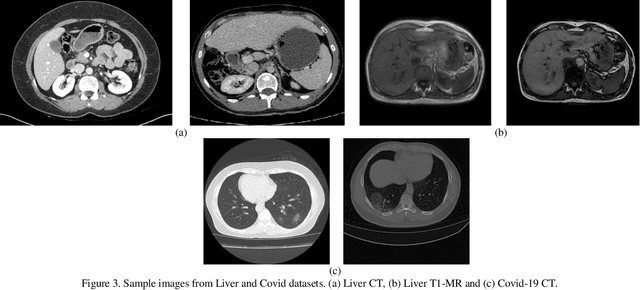
Abstract:Despite the widespread use of deep learning methods for semantic segmentation of images that are acquired from a single source, clinicians often use multi-domain data for a detailed analysis. For instance, CT and MRI have advantages over each other in terms of imaging quality, artifacts, and output characteristics that lead to differential diagnosis. The capacity of current segmentation techniques is only allow to work for an individual domain due to their differences. However, the models that are capable of working on all modalities are essentially needed for a complete solution. Furthermore, robustness is drastically affected by the number of samples in the training step, especially for deep learning models. Hence, there is a necessity that all available data regardless of data domain should be used for reliable methods. For this purpose, this manuscript aims to implement a novel model that can learn robust representations from cross-domain data by encapsulating distinct and shared patterns from different modalities. Precisely, covariate shift property is retained with structural modification and adversarial loss where sparse and rich representations are obtained. Hence, a single parameter set is used to perform cross-domain segmentation task. The superiority of the proposed method is that no information related to modalities are provided in either training or inference phase. The tests on CT and MRI liver data acquired in routine clinical workflows show that the proposed model outperforms all other baseline with a large margin. Experiments are also conducted on Covid-19 dataset that it consists of CT data where significant intra-class visual differences are observed. Similarly, the proposed method achieves the best performance.
CHAOS Challenge -- Combined (CT-MR) Healthy Abdominal Organ Segmentation
Jan 17, 2020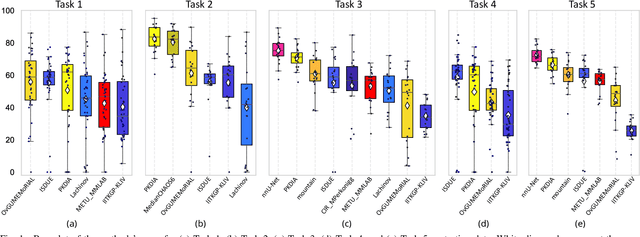
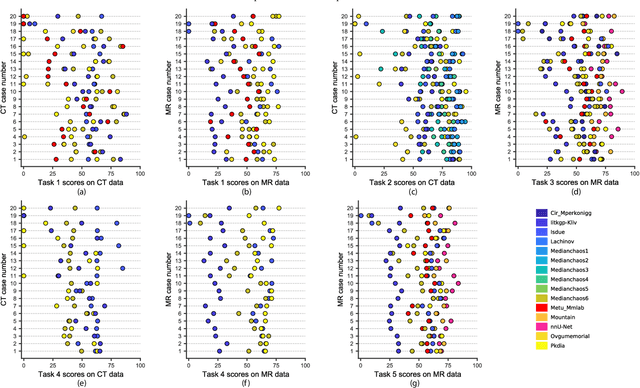
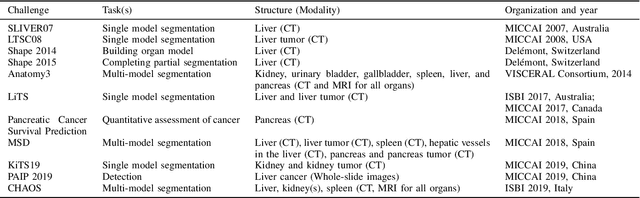
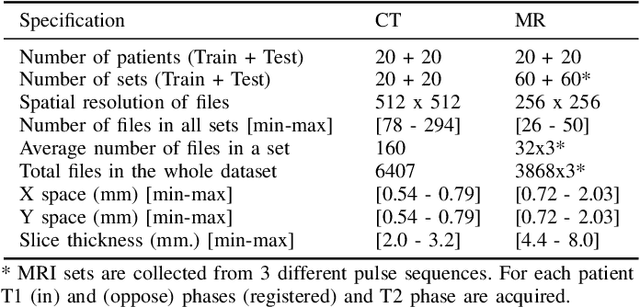
Abstract:Segmentation of abdominal organs has been a comprehensive, yet unresolved, research field for many years. In the last decade, intensive developments in deep learning (DL) have introduced new state-of-the-art segmentation systems. Despite outperforming the overall accuracy of existing systems, the effects of DL model properties and parameters on the performance is hard to interpret. This makes comparative analysis a necessary tool to achieve explainable studies and systems. Moreover, the performance of DL for emerging learning approaches such as cross-modality and multi-modal tasks have been rarely discussed. In order to expand the knowledge in these topics, CHAOS -- Combined (CT-MR) Healthy Abdominal Organ Segmentation challenge has been organized in the IEEE International Symposium on Biomedical Imaging (ISBI), 2019, in Venice, Italy. Despite a large number of the previous abdomen related challenges, the majority of which are focused on tumor/lesion detection and/or classification with a single modality, CHAOS provides both abdominal CT and MR data from healthy subjects. Five different and complementary tasks have been designed to analyze the capabilities of the current approaches from multiple perspectives. The results are investigated thoroughly, compared with manual annotations and interactive methods. The outcomes are reported in detail to reflect the latest advancements in the field. CHAOS challenge and data will be available online to provide a continuous benchmark resource for segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge