Matthew M Churpek
Progress Note Understanding -- Assessment and Plan Reasoning: Overview of the 2022 N2C2 Track 3 Shared Task
Mar 14, 2023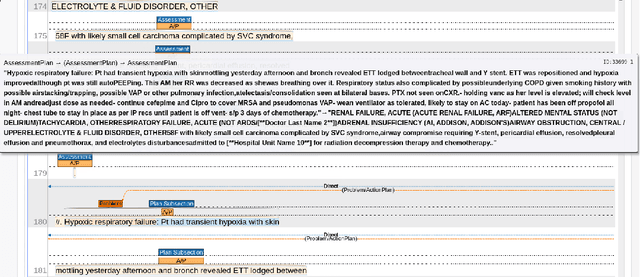
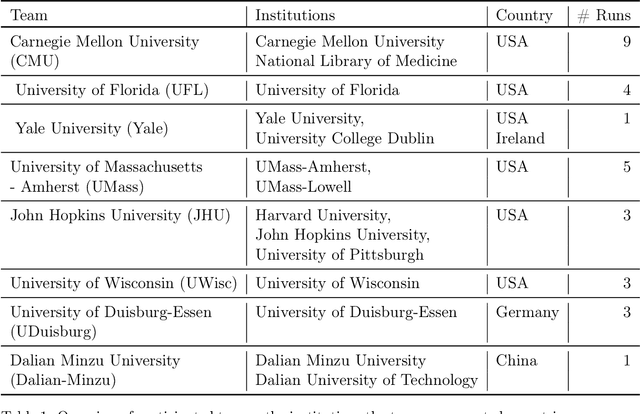
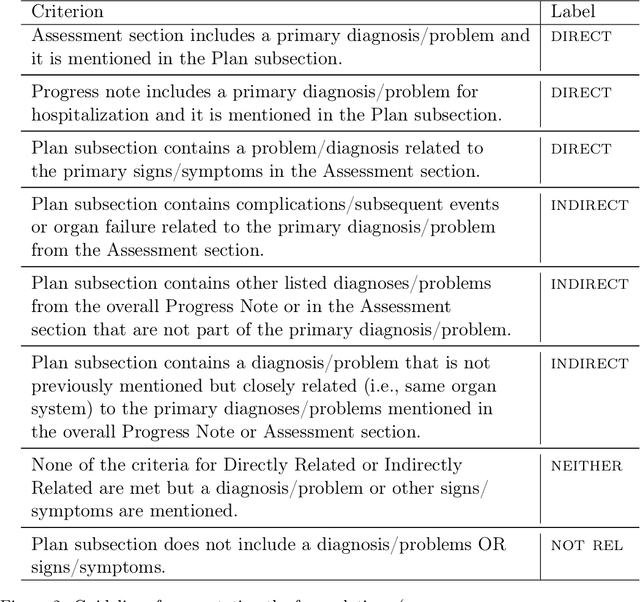
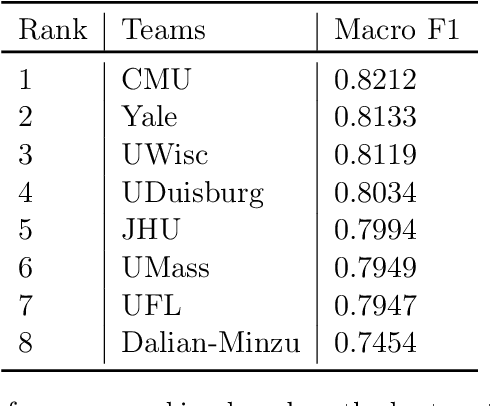
Abstract:Daily progress notes are common types in the electronic health record (EHR) where healthcare providers document the patient's daily progress and treatment plans. The EHR is designed to document all the care provided to patients, but it also enables note bloat with extraneous information that distracts from the diagnoses and treatment plans. Applications of natural language processing (NLP) in the EHR is a growing field with the majority of methods in information extraction. Few tasks use NLP methods for downstream diagnostic decision support. We introduced the 2022 National NLP Clinical Challenge (N2C2) Track 3: Progress Note Understanding - Assessment and Plan Reasoning as one step towards a new suite of tasks. The Assessment and Plan Reasoning task focuses on the most critical components of progress notes, Assessment and Plan subsections where health problems and diagnoses are contained. The goal of the task was to develop and evaluate NLP systems that automatically predict causal relations between the overall status of the patient contained in the Assessment section and its relation to each component of the Plan section which contains the diagnoses and treatment plans. The goal of the task was to identify and prioritize diagnoses as the first steps in diagnostic decision support to find the most relevant information in long documents like daily progress notes. We present the results of 2022 n2c2 Track 3 and provide a description of the data, evaluation, participation and system performance.
DR.BENCH: Diagnostic Reasoning Benchmark for Clinical Natural Language Processing
Sep 29, 2022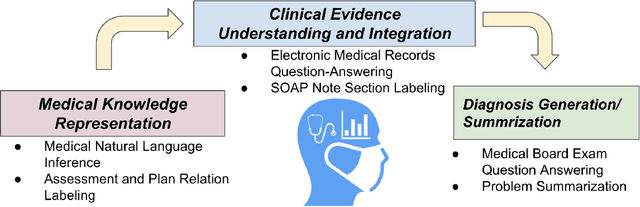
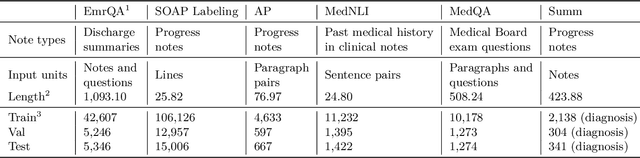
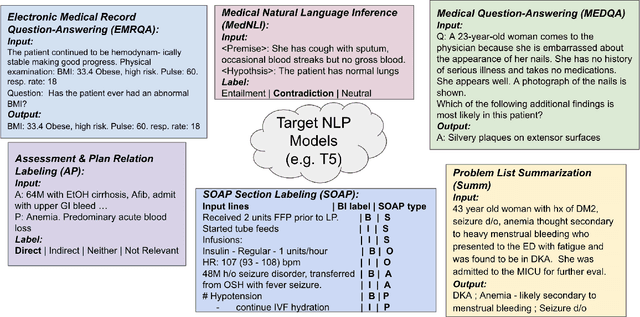
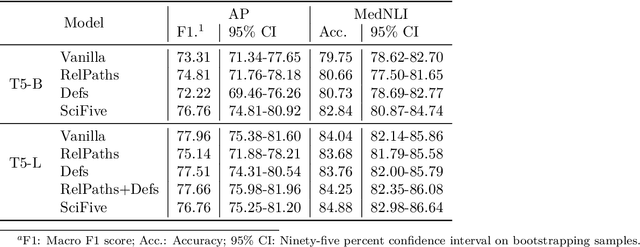
Abstract:The meaningful use of electronic health records (EHR) continues to progress in the digital era with clinical decision support systems augmented by artificial intelligence. A priority in improving provider experience is to overcome information overload and reduce the cognitive burden so fewer medical errors and cognitive biases are introduced during patient care. One major type of medical error is diagnostic error due to systematic or predictable errors in judgment that rely on heuristics. The potential for clinical natural language processing (cNLP) to model diagnostic reasoning in humans with forward reasoning from data to diagnosis and potentially reduce the cognitive burden and medical error has not been investigated. Existing tasks to advance the science in cNLP have largely focused on information extraction and named entity recognition through classification tasks. We introduce a novel suite of tasks coined as Diagnostic Reasoning Benchmarks, DR.BENCH, as a new benchmark for developing and evaluating cNLP models with clinical diagnostic reasoning ability. The suite includes six tasks from ten publicly available datasets addressing clinical text understanding, medical knowledge reasoning, and diagnosis generation. DR.BENCH is the first clinical suite of tasks designed to be a natural language generation framework to evaluate pre-trained language models. Experiments with state-of-the-art pre-trained generative language models using large general domain models and models that were continually trained on a medical corpus demonstrate opportunities for improvement when evaluated in DR. BENCH. We share DR. BENCH as a publicly available GitLab repository with a systematic approach to load and evaluate models for the cNLP community.
A Scoping Review of Publicly Available Language Tasks in Clinical Natural Language Processing
Dec 07, 2021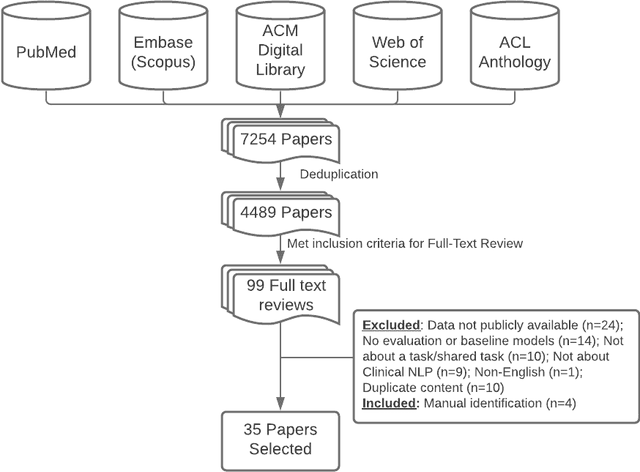
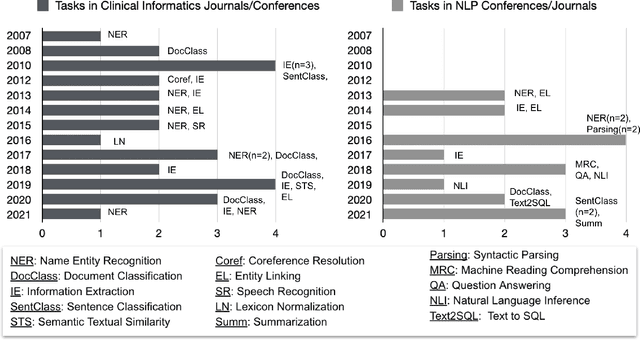

Abstract:Objective: to provide a scoping review of papers on clinical natural language processing (NLP) tasks that use publicly available electronic health record data from a cohort of patients. Materials and Methods: We searched six databases, including biomedical research and computer science literature database. A round of title/abstract screening and full-text screening were conducted by two reviewers. Our method followed the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines. Results: A total of 35 papers with 47 clinical NLP tasks met inclusion criteria between 2007 and 2021. We categorized the tasks by the type of NLP problems, including name entity recognition, summarization, and other NLP tasks. Some tasks were introduced with a topic of clinical decision support applications, such as substance abuse, phenotyping, cohort selection for clinical trial. We summarized the tasks by publication and dataset information. Discussion: The breadth of clinical NLP tasks keeps growing as the field of NLP evolves with advancements in language systems. However, gaps exist in divergent interests between general domain NLP community and clinical informatics community, and in generalizability of the data sources. We also identified issues in data selection and preparation including the lack of time-sensitive data, and invalidity of problem size and evaluation. Conclusions: The existing clinical NLP tasks cover a wide range of topics and the field will continue to grow and attract more attention from both general domain NLP and clinical informatics community. We encourage future work to incorporate multi-disciplinary collaboration, reporting transparency, and standardization in data preparation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge