Matteo Maspero
Quantitative mapping from conventional MRI using self-supervised physics-guided deep learning: applications to a large-scale, clinically heterogeneous dataset
Jan 08, 2026Abstract:Magnetic resonance imaging (MRI) is a cornerstone of clinical neuroimaging, yet conventional MRIs provide qualitative information heavily dependent on scanner hardware and acquisition settings. While quantitative MRI (qMRI) offers intrinsic tissue parameters, the requirement for specialized acquisition protocols and reconstruction algorithms restricts its availability and impedes large-scale biomarker research. This study presents a self-supervised physics-guided deep learning framework to infer quantitative T1, T2, and proton-density (PD) maps directly from widely available clinical conventional T1-weighted, T2-weighted, and FLAIR MRIs. The framework was trained and evaluated on a large-scale, clinically heterogeneous dataset comprising 4,121 scan sessions acquired at our institution over six years on four different 3 T MRI scanner systems, capturing real-world clinical variability. The framework integrates Bloch-based signal models directly into the training objective. Across more than 600 test sessions, the generated maps exhibited white matter and gray matter values consistent with literature ranges. Additionally, the generated maps showed invariance to scanner hardware and acquisition protocol groups, with inter-group coefficients of variation $\leq$ 1.1%. Subject-specific analyses demonstrated excellent voxel-wise reproducibility across scanner systems and sequence parameters, with Pearson $r$ and concordance correlation coefficients exceeding 0.82 for T1 and T2. Mean relative voxel-wise differences were low across all quantitative parameters, especially for T2 ($<$ 6%). These results indicate that the proposed framework can robustly transform diverse clinical conventional MRI data into quantitative maps, potentially paving the way for large-scale quantitative biomarker research.
TrackRAD2025 challenge dataset: Real-time tumor tracking for MRI-guided radiotherapy
Mar 24, 2025Abstract:Purpose: Magnetic resonance imaging (MRI) to visualize anatomical motion is becoming increasingly important when treating cancer patients with radiotherapy. Hybrid MRI-linear accelerator (MRI-linac) systems allow real-time motion management during irradiation. This paper presents a multi-institutional real-time MRI time series dataset from different MRI-linac vendors. The dataset is designed to support developing and evaluating real-time tumor localization (tracking) algorithms for MRI-guided radiotherapy within the TrackRAD2025 challenge (https://trackrad2025.grand-challenge.org/). Acquisition and validation methods: The dataset consists of sagittal 2D cine MRIs in 585 patients from six centers (3 Dutch, 1 German, 1 Australian, and 1 Chinese). Tumors in the thorax, abdomen, and pelvis acquired on two commercially available MRI-linacs (0.35 T and 1.5 T) were included. For 108 cases, irradiation targets or tracking surrogates were manually segmented on each temporal frame. The dataset was randomly split into a public training set of 527 cases (477 unlabeled and 50 labeled) and a private testing set of 58 cases (all labeled). Data Format and Usage Notes: The data is publicly available under the TrackRAD2025 collection: https://doi.org/10.57967/hf/4539. Both the images and segmentations for each patient are available in metadata format. Potential Applications: This novel clinical dataset will enable the development and evaluation of real-time tumor localization algorithms for MRI-guided radiotherapy. By enabling more accurate motion management and adaptive treatment strategies, this dataset has the potential to advance the field of radiotherapy significantly.
SynthRAD2025 Grand Challenge dataset: generating synthetic CTs for radiotherapy
Feb 24, 2025Abstract:Medical imaging is essential in modern radiotherapy, supporting diagnosis, treatment planning, and monitoring. Synthetic imaging, particularly synthetic computed tomography (sCT), is gaining traction in radiotherapy. The SynthRAD2025 dataset and Grand Challenge promote advancements in sCT generation by providing a benchmarking platform for algorithms using cone-beam CT (CBCT) and magnetic resonance imaging (MRI). The dataset includes 2362 cases: 890 MRI-CT and 1472 CBCT-CT pairs from head-and-neck, thoracic, and abdominal cancer patients treated at five European university medical centers (UMC Groningen, UMC Utrecht, Radboud UMC, LMU University Hospital Munich, and University Hospital of Cologne). Data were acquired with diverse scanners and protocols. Pre-processing, including rigid and deformable image registration, ensures high-quality, modality-aligned images. Extensive quality assurance validates image consistency and usability. All imaging data is provided in MetaImage (.mha) format, ensuring compatibility with medical image processing tools. Metadata, including acquisition parameters and registration details, is available in structured CSV files. To maintain dataset integrity, SynthRAD2025 is divided into training (65%), validation (10%), and test (25%) sets. The dataset is accessible at https://doi.org/10.5281/zenodo.14918089 under the SynthRAD2025 collection. This dataset supports benchmarking and the development of synthetic imaging techniques for radiotherapy applications. Use cases include sCT generation for MRI-only and MR-guided photon/proton therapy, CBCT-based dose calculations, and adaptive radiotherapy workflows. By integrating diverse acquisition settings, SynthRAD2025 fosters robust, generalizable image synthesis algorithms, advancing personalized cancer care and adaptive radiotherapy.
Deep learning-based auto-contouring of organs/structures-at-risk for pediatric upper abdominal radiotherapy
Nov 01, 2024
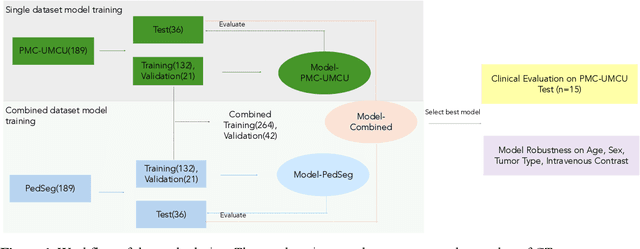


Abstract:Purposes: This study aimed to develop a computed tomography (CT)-based multi-organ segmentation model for delineating organs-at-risk (OARs) in pediatric upper abdominal tumors and evaluate its robustness across multiple datasets. Materials and methods: In-house postoperative CTs from pediatric patients with renal tumors and neuroblastoma (n=189) and a public dataset (n=189) with CTs covering thoracoabdominal regions were used. Seventeen OARs were delineated: nine by clinicians (Type 1) and eight using TotalSegmentator (Type 2). Auto-segmentation models were trained using in-house (ModelPMC-UMCU) and a combined dataset of public data (Model-Combined). Performance was assessed with Dice Similarity Coefficient (DSC), 95% Hausdorff Distance (HD95), and mean surface distance (MSD). Two clinicians rated clinical acceptability on a 5-point Likert scale across 15 patient contours. Model robustness was evaluated against sex, age, intravenous contrast, and tumor type. Results: Model-PMC-UMCU achieved mean DSC values above 0.95 for five of nine OARs, while spleen and heart ranged between 0.90 and 0.95. The stomach-bowel and pancreas exhibited DSC values below 0.90. Model-Combined demonstrated improved robustness across both datasets. Clinical evaluation revealed good usability, with both clinicians rating six of nine Type 1 OARs above four and six of eight Type 2 OARs above three. Significant performance 2 differences were only found across age groups in both datasets, specifically in the left lung and pancreas. The 0-2 age group showed the lowest performance. Conclusion: A multi-organ segmentation model was developed, showcasing enhanced robustness when trained on combined datasets. This model is suitable for various OARs and can be applied to multiple datasets in clinical settings.
Deep learning-based brain segmentation model performance validation with clinical radiotherapy CT
Jun 25, 2024



Abstract:Manual segmentation of medical images is labor intensive and especially challenging for images with poor contrast or resolution. The presence of disease exacerbates this further, increasing the need for an automated solution. To this extent, SynthSeg is a robust deep learning model designed for automatic brain segmentation across various contrasts and resolutions. This study validates the SynthSeg robust brain segmentation model on computed tomography (CT), using a multi-center dataset. An open access dataset of 260 paired CT and magnetic resonance imaging (MRI) from radiotherapy patients treated in 5 centers was collected. Brain segmentations from CT and MRI were obtained with SynthSeg model, a component of the Freesurfer imaging suite. These segmentations were compared and evaluated using Dice scores and Hausdorff 95 distance (HD95), treating MRI-based segmentations as the ground truth. Brain regions that failed to meet performance criteria were excluded based on automated quality control (QC) scores. Dice scores indicate a median overlap of 0.76 (IQR: 0.65-0.83). The median HD95 is 2.95 mm (IQR: 1.73-5.39). QC score based thresholding improves median dice by 0.1 and median HD95 by 0.05mm. Morphological differences related to sex and age, as detected by MRI, were also replicated with CT, with an approximate 17% difference between the CT and MRI results for sex and 10% difference between the results for age. SynthSeg can be utilized for CT-based automatic brain segmentation, but only in applications where precision is not essential. CT performance is lower than MRI based on the integrated QC scores, but low-quality segmentations can be excluded with QC-based thresholding. Additionally, performing CT-based neuroanatomical studies is encouraged, as the results show correlations in sex- and age-based analyses similar to those found with MRI.
Generalizable synthetic MRI with physics-informed convolutional networks
May 21, 2023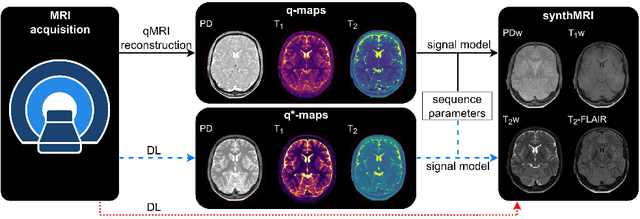

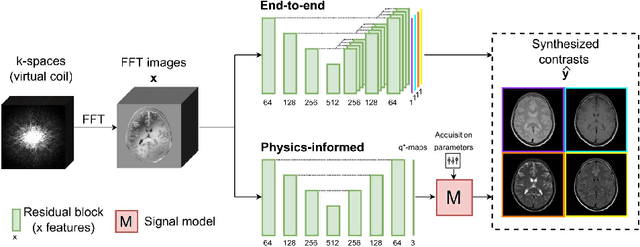

Abstract:In this study, we develop a physics-informed deep learning-based method to synthesize multiple brain magnetic resonance imaging (MRI) contrasts from a single five-minute acquisition and investigate its ability to generalize to arbitrary contrasts to accelerate neuroimaging protocols. A dataset of fifty-five subjects acquired with a standard MRI protocol and a five-minute transient-state sequence was used to develop a physics-informed deep learning-based method. The model, based on a generative adversarial network, maps data acquired from the five-minute scan to "effective" quantitative parameter maps, here named q*-maps, by using its generated PD, T1, and T2 values in a signal model to synthesize four standard contrasts (proton density-weighted, T1-weighted, T2-weighted, and T2-weighted fluid-attenuated inversion recovery), from which losses are computed. The q*-maps are compared to literature values and the synthetic contrasts are compared to an end-to-end deep learning-based method proposed by literature. The generalizability of the proposed method is investigated for five volunteers by synthesizing three non-standard contrasts unseen during training and comparing these to respective ground truth acquisitions via contrast-to-noise ratio and quantitative assessment. The physics-informed method was able to match the high-quality synthMRI of the end-to-end method for the four standard contrasts, with mean \pm standard deviation structural similarity metrics above 0.75 \pm 0.08 and peak signal-to-noise ratios above 22.4 \pm 1.9 and 22.6 \pm 2.1. Additionally, the physics-informed method provided retrospective contrast adjustment, with visually similar signal contrast and comparable contrast-to-noise ratios to the ground truth acquisitions for three sequences unused for model training, demonstrating its generalizability and potential application to accelerate neuroimaging protocols.
SynthRAD2023 Grand Challenge dataset: generating synthetic CT for radiotherapy
Mar 28, 2023



Abstract:Purpose: Medical imaging has become increasingly important in diagnosing and treating oncological patients, particularly in radiotherapy. Recent advances in synthetic computed tomography (sCT) generation have increased interest in public challenges to provide data and evaluation metrics for comparing different approaches openly. This paper describes a dataset of brain and pelvis computed tomography (CT) images with rigidly registered CBCT and MRI images to facilitate the development and evaluation of sCT generation for radiotherapy planning. Acquisition and validation methods: The dataset consists of CT, CBCT, and MRI of 540 brains and 540 pelvic radiotherapy patients from three Dutch university medical centers. Subjects' ages ranged from 3 to 93 years, with a mean age of 60. Various scanner models and acquisition settings were used across patients from the three data-providing centers. Details are available in CSV files provided with the datasets. Data format and usage notes: The data is available on Zenodo (https://doi.org/10.5281/zenodo.7260705) under the SynthRAD2023 collection. The images for each subject are available in nifti format. Potential applications: This dataset will enable the evaluation and development of image synthesis algorithms for radiotherapy purposes on a realistic multi-center dataset with varying acquisition protocols. Synthetic CT generation has numerous applications in radiation therapy, including diagnosis, treatment planning, treatment monitoring, and surgical planning.
Exploring contrast generalisation in deep learning-based brain MRI-to-CT synthesis
Mar 17, 2023



Abstract:Background: Synthetic computed tomography (sCT) has been proposed and increasingly clinically adopted to enable magnetic resonance imaging (MRI)-based radiotherapy. Deep learning (DL) has recently demonstrated the ability to generate accurate sCT from fixed MRI acquisitions. However, MRI protocols may change over time or differ between centres resulting in low-quality sCT due to poor model generalisation. Purpose: investigating domain randomisation (DR) to increase the generalisation of a DL model for brain sCT generation. Methods: CT and corresponding T1-weighted MRI with/without contrast, T2-weighted, and FLAIR MRI from 95 patients undergoing RT were collected, considering FLAIR the unseen sequence where to investigate generalisation. A ``Baseline'' generative adversarial network was trained with/without the FLAIR sequence to test how a model performs without DR. Image similarity and accuracy of sCT-based dose plans were assessed against CT to select the best-performing DR approach against the Baseline. Results: The Baseline model had the poorest performance on FLAIR, with mean absolute error (MAE)=106$\pm$20.7 HU (mean$\pm\sigma$). Performance on FLAIR significantly improved for the DR model with MAE=99.0$\pm$14.9 HU, but still inferior to the performance of the Baseline+FLAIR model (MAE=72.6$\pm$10.1 HU). Similarly, an improvement in $\gamma$-pass rate was obtained for DR vs Baseline. Conclusions: DR improved image similarity and dose accuracy on the unseen sequence compared to training only on acquired MRI. DR makes the model more robust, reducing the need for re-training when applying a model on sequences unseen and unavailable for retraining.
Accelerated respiratory-resolved 4D-MRI with separable spatio-temporal neural networks
Nov 10, 2022Abstract:Purpose: To quickly obtain high-quality respiratory-resolved four-dimensional magnetic resonance imaging (4D-MRI), enabling accurate motion quantification for MRI-guided radiotherapy. Methods: A small convolutional neural network called MODEST is proposed to reconstruct 4D-MRI by performing a spatial and temporal decomposition, omitting the need for 4D convolutions to use all the spatio-temporal information present in 4D-MRI. This network is trained on undersampled 4D-MRI after respiratory binning to reconstruct high-quality 4D-MRI obtained by compressed sensing reconstruction. The network is trained, validated, and tested on 4D-MRI of 28 lung cancer patients acquired with a T1-weighted golden-angle radial stack-of-stars sequence. The 4D-MRI of 18, 5, and 5 patients were used for training, validation, and testing. Network performances are evaluated on image quality measured by the structural similarity index (SSIM) and motion consistency by comparing the position of the lung-liver interface on undersampled 4D-MRI before and after respiratory binning. The network is compared to conventional architectures such as a U-Net, which has 30 times more trainable parameters. Results: MODEST can reconstruct high-quality 4D-MRI with higher image quality than a U-Net, despite a thirty-fold reduction in trainable parameters. High-quality 4D-MRI can be obtained using MODEST in approximately 2.5 minutes, including acquisition, processing, and reconstruction. Conclusion: High-quality accelerated 4D-MRI can be obtained using MODEST, which is particularly interesting for MRI-guided radiotherapy.
Deep learning-based synthetic-CT generation in radiotherapy and PET: a review
Feb 04, 2021



Abstract:Recently, deep learning (DL)-based methods for the generation of synthetic computed tomography (sCT) have received significant research attention as an alternative to classical ones. We present here a systematic review of these methods by grouping them into three categories, according to their clinical applications: I) to replace CT in magnetic resonance (MR)-based treatment planning, II) facilitate cone-beam computed tomography (CBCT)-based image-guided adaptive radiotherapy, and III) derive attenuation maps for the correction of Positron Emission Tomography (PET). Appropriate database searching was performed on journal articles published between January 2014 and December 2020. The DL methods' key characteristics were extracted from each eligible study, and a comprehensive comparison among network architectures and metrics was reported. A detailed review of each category was given, highlighting essential contributions, identifying specific challenges, and summarising the achievements. Lastly, the statistics of all the cited works from various aspects were analysed, revealing the popularity and future trends, and the potential of DL-based sCT generation. The current status of DL-based sCT generation was evaluated, assessing the clinical readiness of the presented methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge