Marylyn D. Ritchie
Leveraging Social Determinants of Health in Alzheimer's Research Using LLM-Augmented Literature Mining and Knowledge Graphs
Oct 04, 2024Abstract:Growing evidence suggests that social determinants of health (SDoH), a set of nonmedical factors, affect individuals' risks of developing Alzheimer's disease (AD) and related dementias. Nevertheless, the etiological mechanisms underlying such relationships remain largely unclear, mainly due to difficulties in collecting relevant information. This study presents a novel, automated framework that leverages recent advancements of large language model (LLM) and natural language processing techniques to mine SDoH knowledge from extensive literature and integrate it with AD-related biological entities extracted from the general-purpose knowledge graph PrimeKG. Utilizing graph neural networks, we performed link prediction tasks to evaluate the resultant SDoH-augmented knowledge graph. Our framework shows promise for enhancing knowledge discovery in AD and can be generalized to other SDoH-related research areas, offering a new tool for exploring the impact of social determinants on health outcomes. Our code is available at: https://github.com/hwq0726/SDoHenPKG
Multidimensional representations in late-life depression: convergence in neuroimaging, cognition, clinical symptomatology and genetics
Oct 25, 2021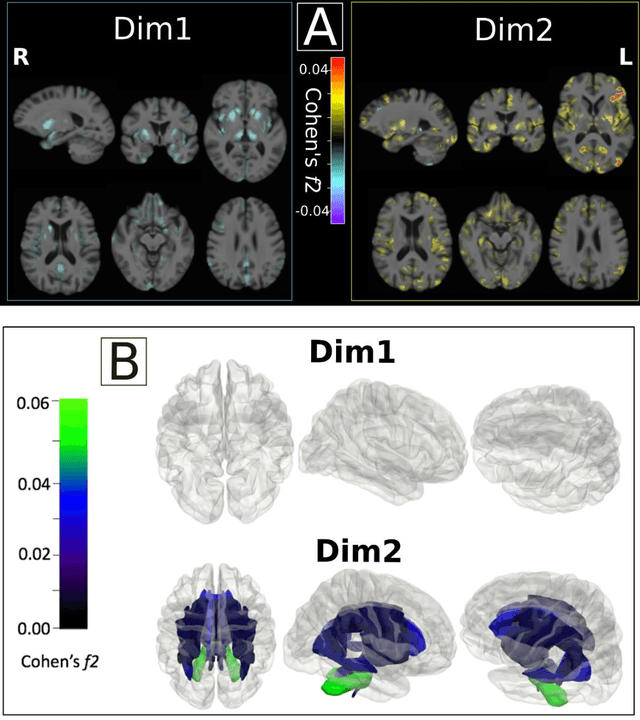
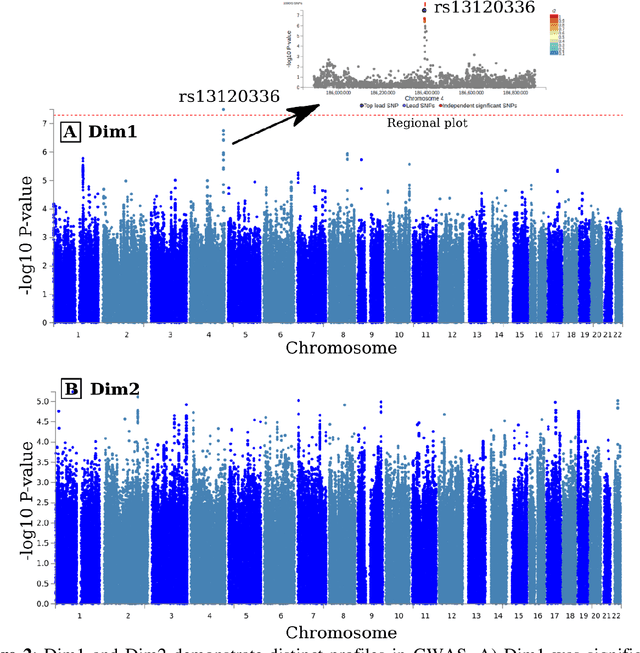
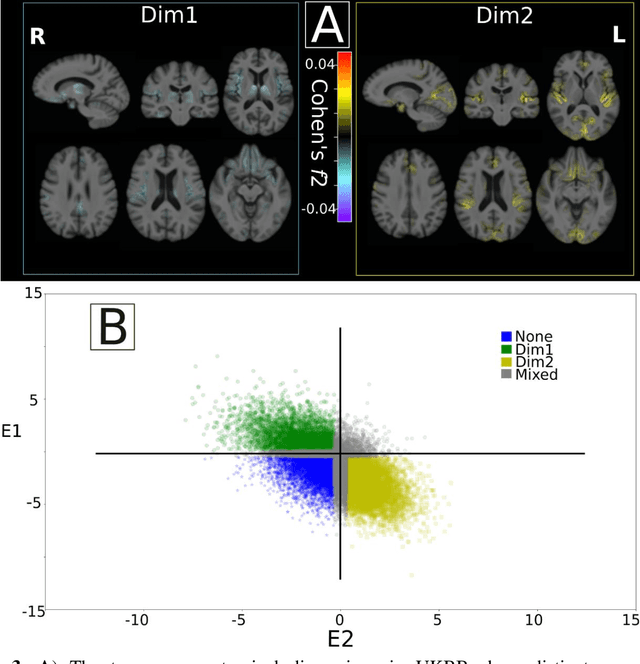
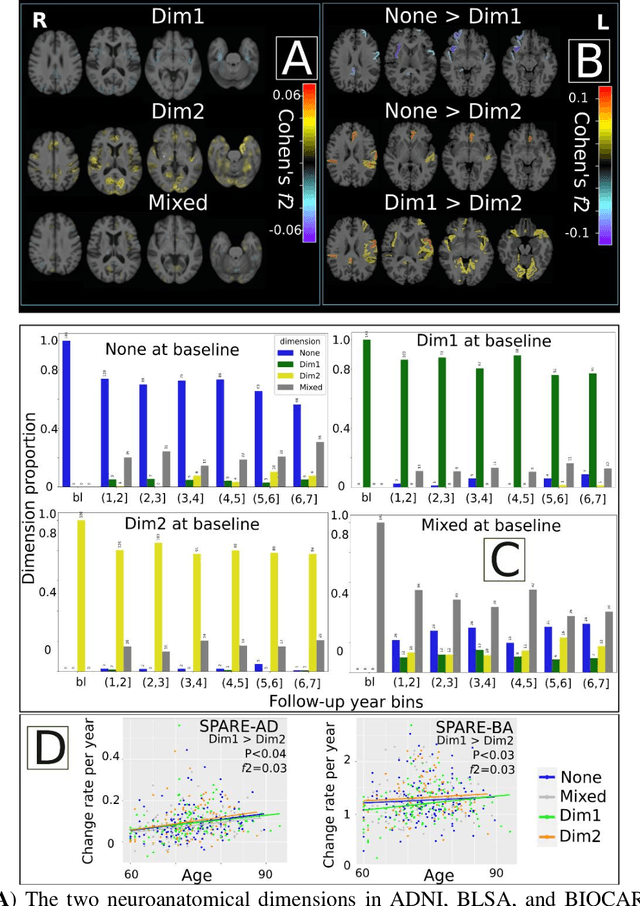
Abstract:Late-life depression (LLD) is characterized by considerable heterogeneity in clinical manifestation. Unraveling such heterogeneity would aid in elucidating etiological mechanisms and pave the road to precision and individualized medicine. We sought to delineate, cross-sectionally and longitudinally, disease-related heterogeneity in LLD linked to neuroanatomy, cognitive functioning, clinical symptomatology, and genetic profiles. Multimodal data from a multicentre sample (N=996) were analyzed. A semi-supervised clustering method (HYDRA) was applied to regional grey matter (GM) brain volumes to derive dimensional representations. Two dimensions were identified, which accounted for the LLD-related heterogeneity in voxel-wise GM maps, white matter (WM) fractional anisotropy (FA), neurocognitive functioning, clinical phenotype, and genetics. Dimension one (Dim1) demonstrated relatively preserved brain anatomy without WM disruptions relative to healthy controls. In contrast, dimension two (Dim2) showed widespread brain atrophy and WM integrity disruptions, along with cognitive impairment and higher depression severity. Moreover, one de novo independent genetic variant (rs13120336) was significantly associated with Dim 1 but not with Dim 2. Notably, the two dimensions demonstrated significant SNP-based heritability of 18-27% within the general population (N=12,518 in UKBB). Lastly, in a subset of individuals having longitudinal measurements, Dim2 demonstrated a more rapid longitudinal decrease in GM and brain age, and was more likely to progress to Alzheimers disease, compared to Dim1 (N=1,413 participants and 7,225 scans from ADNI, BLSA, and BIOCARD datasets).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge