Marc-André Legault
On the Challenges of using Reinforcement Learning in Precision Drug Dosing: Delay and Prolongedness of Action Effects
Jan 02, 2023Abstract:Drug dosing is an important application of AI, which can be formulated as a Reinforcement Learning (RL) problem. In this paper, we identify two major challenges of using RL for drug dosing: delayed and prolonged effects of administering medications, which break the Markov assumption of the RL framework. We focus on prolongedness and define PAE-POMDP (Prolonged Action Effect-Partially Observable Markov Decision Process), a subclass of POMDPs in which the Markov assumption does not hold specifically due to prolonged effects of actions. Motivated by the pharmacology literature, we propose a simple and effective approach to converting drug dosing PAE-POMDPs into MDPs, enabling the use of the existing RL algorithms to solve such problems. We validate the proposed approach on a toy task, and a challenging glucose control task, for which we devise a clinically-inspired reward function. Our results demonstrate that: (1) the proposed method to restore the Markov assumption leads to significant improvements over a vanilla baseline; (2) the approach is competitive with recurrent policies which may inherently capture the prolonged effect of actions; (3) it is remarkably more time and memory efficient than the recurrent baseline and hence more suitable for real-time dosing control systems; and (4) it exhibits favorable qualitative behavior in our policy analysis.
Deep interpretability for GWAS
Jul 03, 2020


Abstract:Genome-Wide Association Studies are typically conducted using linear models to find genetic variants associated with common diseases. In these studies, association testing is done on a variant-by-variant basis, possibly missing out on non-linear interaction effects between variants. Deep networks can be used to model these interactions, but they are difficult to train and interpret on large genetic datasets. We propose a method that uses the gradient based deep interpretability technique named DeepLIFT to show that known diabetes genetic risk factors can be identified using deep models along with possibly novel associations.
Diet Networks: Thin Parameters for Fat Genomics
Mar 16, 2017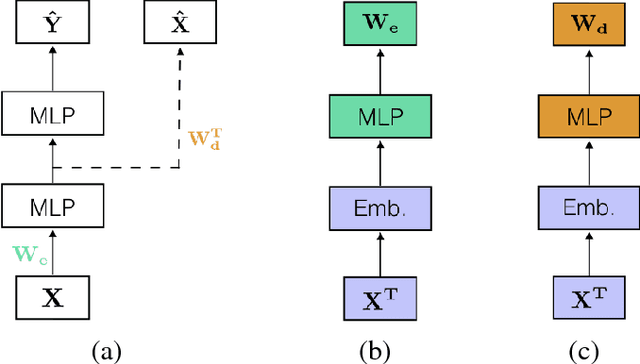
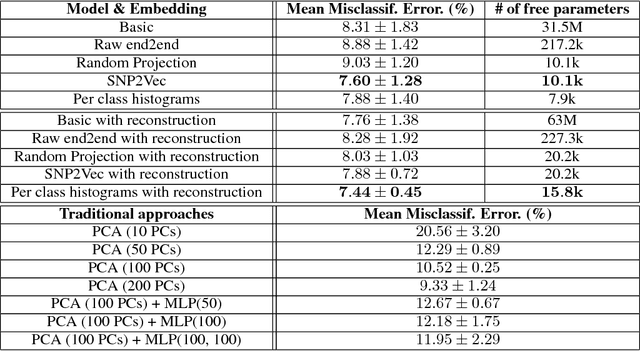
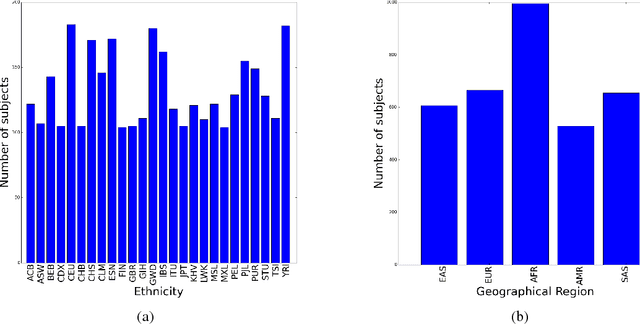
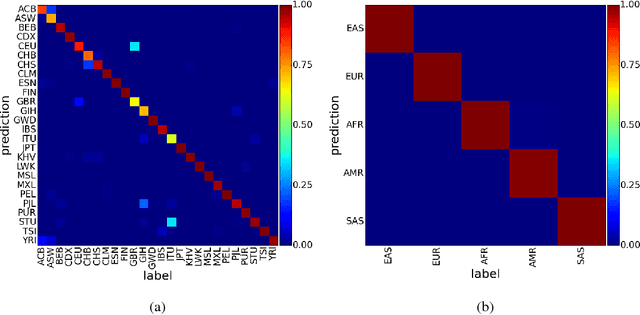
Abstract:Learning tasks such as those involving genomic data often poses a serious challenge: the number of input features can be orders of magnitude larger than the number of training examples, making it difficult to avoid overfitting, even when using the known regularization techniques. We focus here on tasks in which the input is a description of the genetic variation specific to a patient, the single nucleotide polymorphisms (SNPs), yielding millions of ternary inputs. Improving the ability of deep learning to handle such datasets could have an important impact in precision medicine, where high-dimensional data regarding a particular patient is used to make predictions of interest. Even though the amount of data for such tasks is increasing, this mismatch between the number of examples and the number of inputs remains a concern. Naive implementations of classifier neural networks involve a huge number of free parameters in their first layer: each input feature is associated with as many parameters as there are hidden units. We propose a novel neural network parametrization which considerably reduces the number of free parameters. It is based on the idea that we can first learn or provide a distributed representation for each input feature (e.g. for each position in the genome where variations are observed), and then learn (with another neural network called the parameter prediction network) how to map a feature's distributed representation to the vector of parameters specific to that feature in the classifier neural network (the weights which link the value of the feature to each of the hidden units). We show experimentally on a population stratification task of interest to medical studies that the proposed approach can significantly reduce both the number of parameters and the error rate of the classifier.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge