M. N. S. Swamy
Senior Member, IEEE
ECG-EmotionNet: Nested Mixture of Expert (NMoE) Adaptation of ECG-Foundation Model for Driver Emotion Recognition
Mar 03, 2025



Abstract:Driver emotion recognition plays a crucial role in driver monitoring systems, enhancing human-autonomy interactions and the trustworthiness of Autonomous Driving (AD). Various physiological and behavioural modalities have been explored for this purpose, with Electrocardiogram (ECG) emerging as a standout choice for real-time emotion monitoring, particularly in dynamic and unpredictable driving conditions. Existing methods, however, often rely on multi-channel ECG signals recorded under static conditions, limiting their applicability in real-world dynamic driving scenarios. To address this limitation, the paper introduces ECG-EmotionNet, a novel architecture designed specifically for emotion recognition in dynamic driving environments. ECG-EmotionNet is constructed by adapting a recently introduced ECG Foundation Model (FM) and uniquely employs single-channel ECG signals, ensuring both robust generalizability and computational efficiency. Unlike conventional adaptation methods such as full fine-tuning, linear probing, or low-rank adaptation, we propose an intuitively pleasing alternative, referred to as the nested Mixture of Experts (MoE) adaptation. More precisely, each transformer layer of the underlying FM is treated as a separate expert, with embeddings extracted from these experts fused using trainable weights within a gating mechanism. This approach enhances the representation of both global and local ECG features, leading to a 6% improvement in accuracy and a 7% increase in the F1 score, all while maintaining computational efficiency. The effectiveness of the proposed ECG-EmotionNet architecture is evaluated using a recently introduced and challenging driver emotion monitoring dataset.
PathoWAve: A Deep Learning-based Weight Averaging Method for Improving Domain Generalization in Histopathology Images
Jun 21, 2024



Abstract:Recent advancements in deep learning (DL) have significantly advanced medical image analysis. In the field of medical image processing, particularly in histopathology image analysis, the variation in staining protocols and differences in scanners present significant domain shift challenges, undermine the generalization capabilities of models to the data from unseen domains, prompting the need for effective domain generalization (DG) strategies to improve the consistency and reliability of automated cancer detection tools in diagnostic decision-making. In this paper, we introduce Pathology Weight Averaging (PathoWAve), a multi-source DG strategy for addressing domain shift phenomenon of DL models in histopathology image analysis. Integrating specific weight averaging technique with parallel training trajectories and a strategically combination of regular augmentations with histopathology-specific data augmentation methods, PathoWAve enables a comprehensive exploration and precise convergence within the loss landscape. This method significantly enhanced generalization capabilities of DL models across new, unseen histopathology domains. To the best of our knowledge, PathoWAve is the first proposed weight averaging method for DG in histopathology image analysis. Our quantitative results on Camelyon17 WILDS dataset demonstrate PathoWAve's superiority over previous proposed methods to tackle the domain shift phenomenon in histopathology image processing. Our code is available at \url{https://github.com/ParastooSotoudeh/PathoWAve}.
Orthogonal Features-based EEG Signal Denoising using Fractionally Compressed AutoEncoder
Feb 16, 2021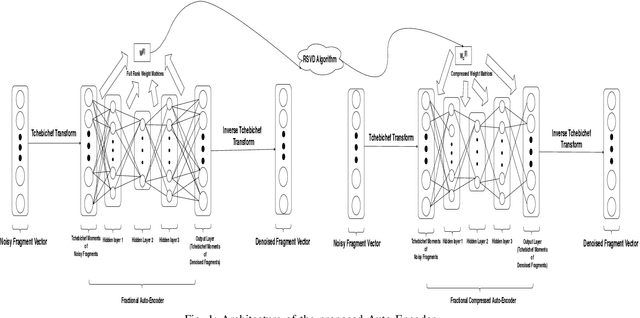
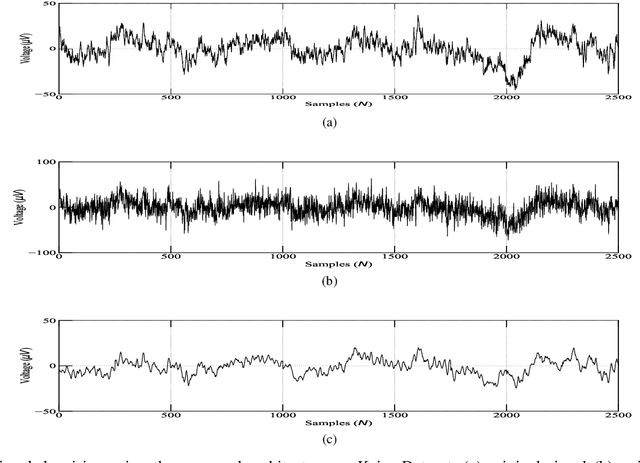
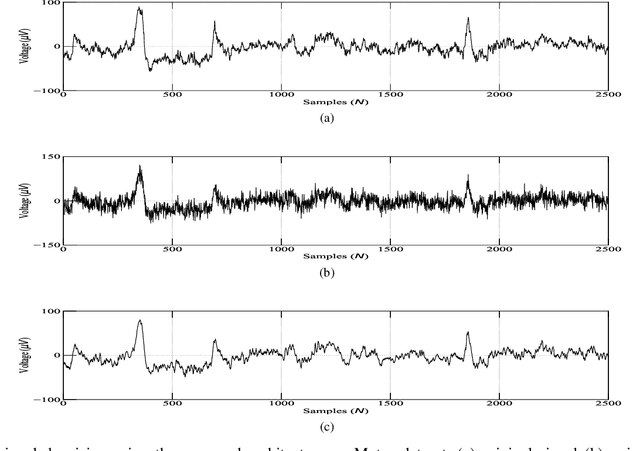
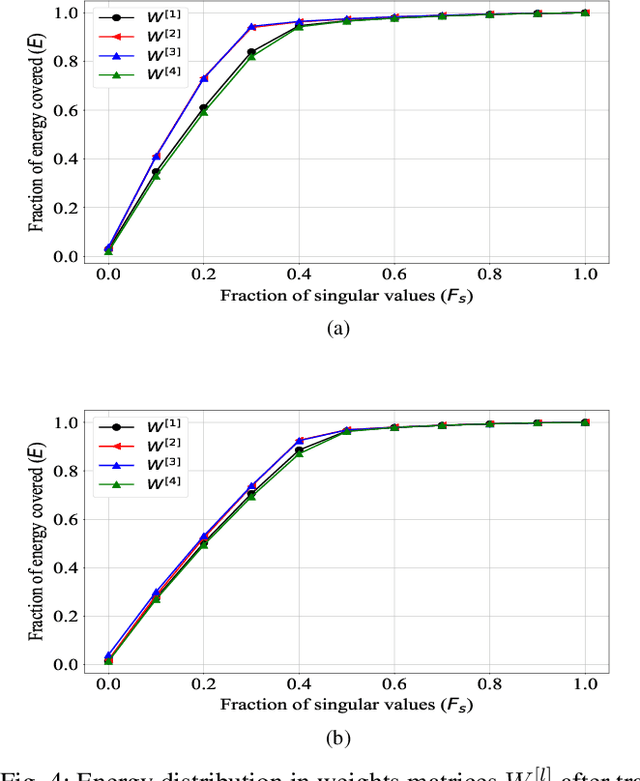
Abstract:A fractional-based compressed auto-encoder architecture has been introduced to solve the problem of denoising electroencephalogram (EEG) signals. The architecture makes use of fractional calculus to calculate the gradients during the backpropagation process, as a result of which a new hyper-parameter in the form of fractional order ($\alpha$) has been introduced which can be tuned to get the best denoising performance. Additionally, to avoid substantial use of memory resources, the model makes use of orthogonal features in the form of Tchebichef moments as input. The orthogonal features have been used in achieving compression at the input stage. Considering the growing use of low energy devices, compression of neural networks becomes imperative. Here, the auto-encoder's weights are compressed using the randomized singular value decomposition (RSVD) algorithm during training while evaluation is performed using various compression ratios. The experimental results show that the proposed fractionally compressed architecture provides improved denoising results on the standard datasets when compared with the existing methods.
Multi-Site Infant Brain Segmentation Algorithms: The iSeg-2019 Challenge
Jul 11, 2020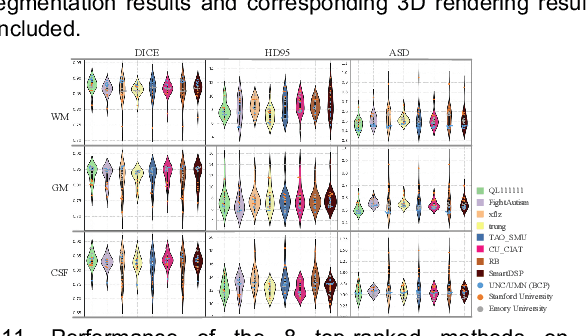
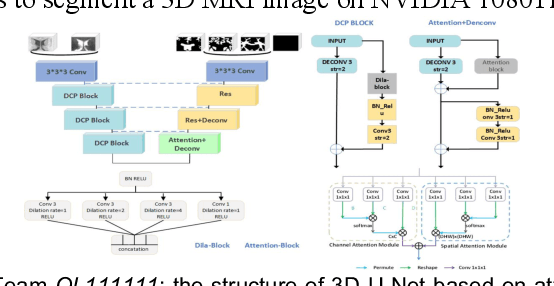
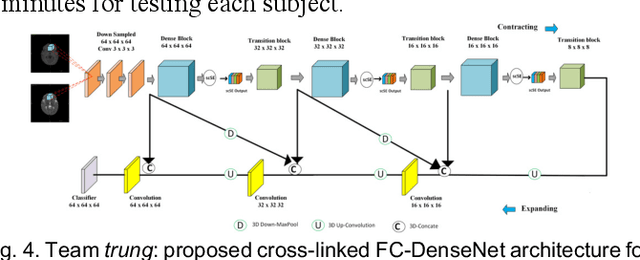
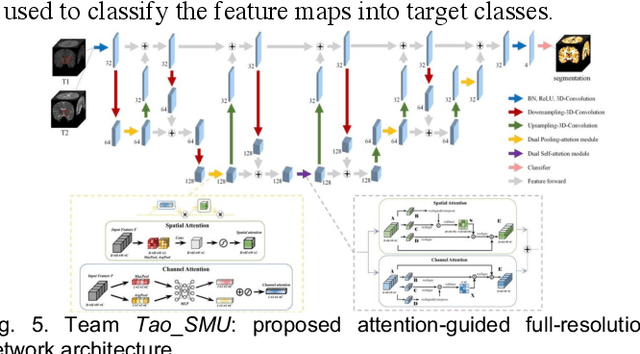
Abstract:To better understand early brain growth patterns in health and disorder, it is critical to accurately segment infant brain magnetic resonance (MR) images into white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF). Deep learning-based methods have achieved state-of-the-art performance; however, one of major limitations is that the learning-based methods may suffer from the multi-site issue, that is, the models trained on a dataset from one site may not be applicable to the datasets acquired from other sites with different imaging protocols/scanners. To promote methodological development in the community, iSeg-2019 challenge (http://iseg2019.web.unc.edu) provides a set of 6-month infant subjects from multiple sites with different protocols/scanners for the participating methods. Training/validation subjects are from UNC (MAP) and testing subjects are from UNC/UMN (BCP), Stanford University, and Emory University. By the time of writing, there are 30 automatic segmentation methods participating in iSeg-2019. We review the 8 top-ranked teams by detailing their pipelines/implementations, presenting experimental results and evaluating performance in terms of the whole brain, regions of interest, and gyral landmark curves. We also discuss their limitations and possible future directions for the multi-site issue. We hope that the multi-site dataset in iSeg-2019 and this review article will attract more researchers on the multi-site issue.
An Efficient Secure Multimodal Biometric Fusion Using Palmprint and Face Image
Sep 12, 2009Abstract:Biometrics based personal identification is regarded as an effective method for automatically recognizing, with a high confidence a person's identity. A multimodal biometric systems consolidate the evidence presented by multiple biometric sources and typically better recognition performance compare to system based on a single biometric modality. This paper proposes an authentication method for a multimodal biometric system identification using two traits i.e. face and palmprint. The proposed system is designed for application where the training data contains a face and palmprint. Integrating the palmprint and face features increases robustness of the person authentication. The final decision is made by fusion at matching score level architecture in which features vectors are created independently for query measures and are then compared to the enrolment template, which are stored during database preparation. Multimodal biometric system is developed through fusion of face and palmprint recognition.
* International Journal of Computer Science Issues (IJCSI), Volume 1, pp49-53, August 2009
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge