Li-ping Liu
NVDiff: Graph Generation through the Diffusion of Node Vectors
Nov 19, 2022



Abstract:Learning to generate graphs is challenging as a graph is a set of pairwise connected, unordered nodes encoding complex combinatorial structures. Recently, several works have proposed graph generative models based on normalizing flows or score-based diffusion models. However, these models need to generate nodes and edges in parallel from the same process, whose dimensionality is unnecessarily high. We propose NVDiff, which takes the VGAE structure and uses a score-based generative model (SGM) as a flexible prior to sample node vectors. By modeling only node vectors in the latent space, NVDiff significantly reduces the dimension of the diffusion process and thus improves sampling speed. Built on the NVDiff framework, we introduce an attention-based score network capable of capturing both local and global contexts of graphs. Experiments indicate that NVDiff significantly reduces computations and can model much larger graphs than competing methods. At the same time, it achieves superior or competitive performances over various datasets compared to previous methods.
Ensemble Spectral Prediction (ESP) Model for Metabolite Annotation
Mar 25, 2022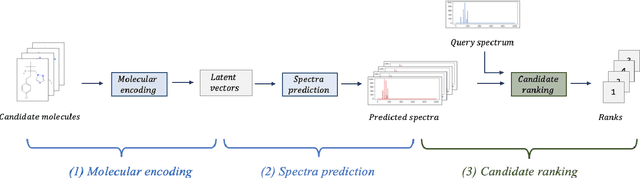
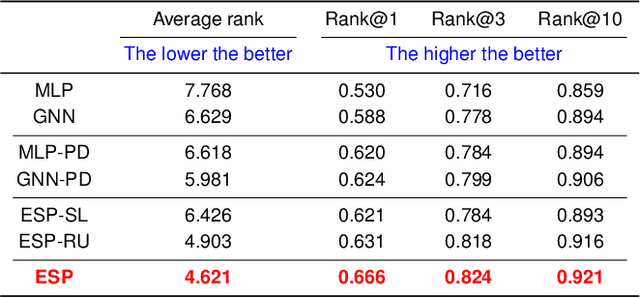
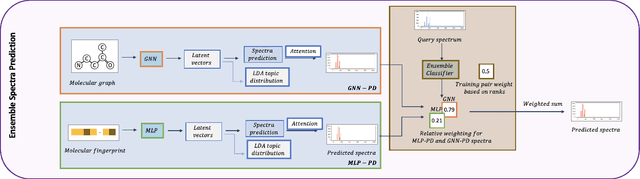
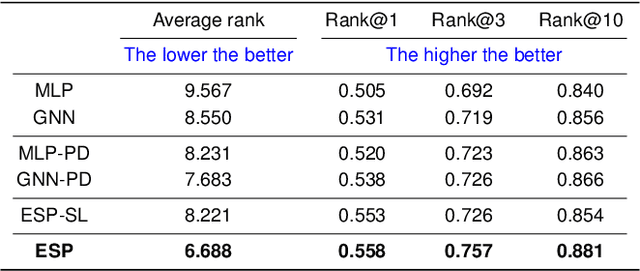
Abstract:A key challenge in metabolomics is annotating measured spectra from a biological sample with chemical identities. Currently, only a small fraction of measurements can be assigned identities. Two complementary computational approaches have emerged to address the annotation problem: mapping candidate molecules to spectra, and mapping query spectra to molecular candidates. In essence, the candidate molecule with the spectrum that best explains the query spectrum is recommended as the target molecule. Despite candidate ranking being fundamental in both approaches, no prior works utilized rank learning tasks in determining the target molecule. We propose a novel machine learning model, Ensemble Spectral Prediction (ESP), for metabolite annotation. ESP takes advantage of prior neural network-based annotation models that utilize multilayer perceptron (MLP) networks and Graph Neural Networks (GNNs). Based on the ranking results of the MLP and GNN-based models, ESP learns a weighting for the outputs of MLP and GNN spectral predictors to generate a spectral prediction for a query molecule. Importantly, training data is stratified by molecular formula to provide candidate sets during model training. Further, baseline MLP and GNN models are enhanced by considering peak dependencies through multi-head attention mechanism and multi-tasking on spectral topic distributions. ESP improves average rank by 41% and 30% over the MLP and GNN baselines, respectively, demonstrating remarkable performance gain over state-of-the-art neural network approaches. We show that annotation performance, for ESP and other models, is a strong function of the number of molecules in the candidate set and their similarity to the target molecule.
Boost-RS: Boosted Embeddings for Recommender Systems and its Application to Enzyme-Substrate Interaction Prediction
Sep 28, 2021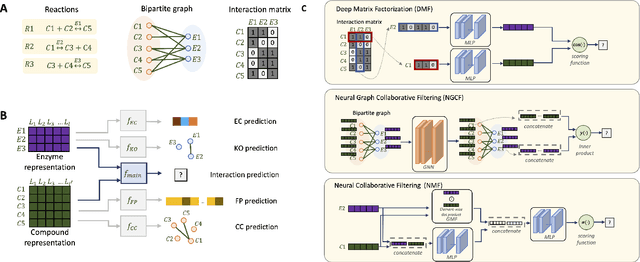
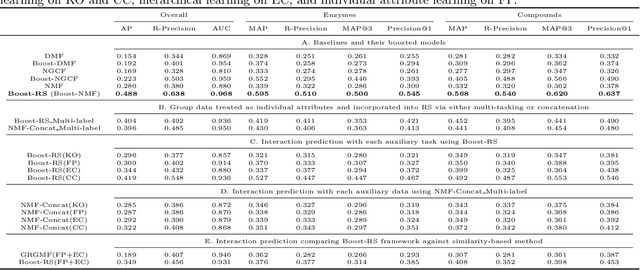
Abstract:Despite experimental and curation efforts, the extent of enzyme promiscuity on substrates continues to be largely unexplored and under documented. Recommender systems (RS), which are currently unexplored for the enzyme-substrate interaction prediction problem, can be utilized to provide enzyme recommendations for substrates, and vice versa. The performance of Collaborative-Filtering (CF) recommender systems however hinges on the quality of embedding vectors of users and items (enzymes and substrates in our case). Importantly, enhancing CF embeddings with heterogeneous auxiliary data, specially relational data (e.g., hierarchical, pairwise, or groupings), remains a challenge. We propose an innovative general RS framework, termed Boost-RS, that enhances RS performance by "boosting" embedding vectors through auxiliary data. Specifically, Boost-RS is trained and dynamically tuned on multiple relevant auxiliary learning tasks Boost-RS utilizes contrastive learning tasks to exploit relational data. To show the efficacy of Boost-RS for the enzyme-substrate prediction interaction problem, we apply the Boost-RS framework to several baseline CF models. We show that each of our auxiliary tasks boosts learning of the embedding vectors, and that contrastive learning using Boost-RS outperforms attribute concatenation and multi-label learning. We also show that Boost-RS outperforms similarity-based models. Ablation studies and visualization of learned representations highlight the importance of using contrastive learning on some of the auxiliary data in boosting the embedding vectors.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge