Ensemble Spectral Prediction (ESP) Model for Metabolite Annotation
Paper and Code
Mar 25, 2022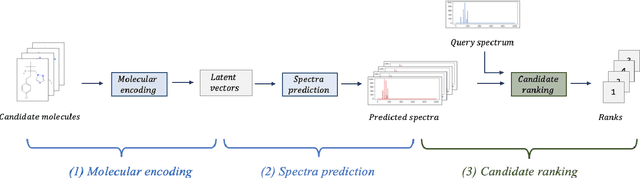
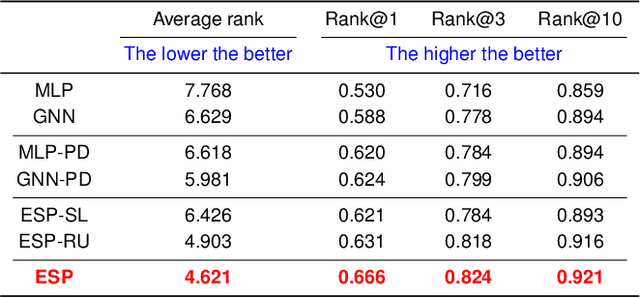
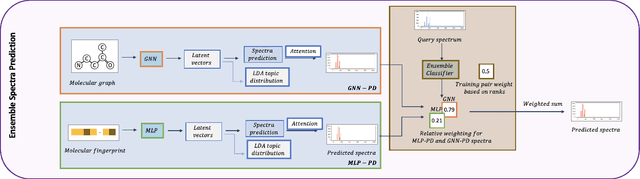
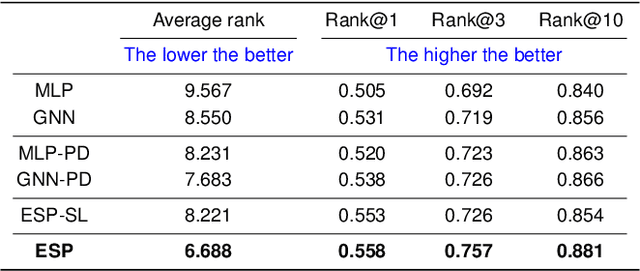
A key challenge in metabolomics is annotating measured spectra from a biological sample with chemical identities. Currently, only a small fraction of measurements can be assigned identities. Two complementary computational approaches have emerged to address the annotation problem: mapping candidate molecules to spectra, and mapping query spectra to molecular candidates. In essence, the candidate molecule with the spectrum that best explains the query spectrum is recommended as the target molecule. Despite candidate ranking being fundamental in both approaches, no prior works utilized rank learning tasks in determining the target molecule. We propose a novel machine learning model, Ensemble Spectral Prediction (ESP), for metabolite annotation. ESP takes advantage of prior neural network-based annotation models that utilize multilayer perceptron (MLP) networks and Graph Neural Networks (GNNs). Based on the ranking results of the MLP and GNN-based models, ESP learns a weighting for the outputs of MLP and GNN spectral predictors to generate a spectral prediction for a query molecule. Importantly, training data is stratified by molecular formula to provide candidate sets during model training. Further, baseline MLP and GNN models are enhanced by considering peak dependencies through multi-head attention mechanism and multi-tasking on spectral topic distributions. ESP improves average rank by 41% and 30% over the MLP and GNN baselines, respectively, demonstrating remarkable performance gain over state-of-the-art neural network approaches. We show that annotation performance, for ESP and other models, is a strong function of the number of molecules in the candidate set and their similarity to the target molecule.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge