Leopold Grinberg
Can a Fruit Fly Learn Word Embeddings?
Jan 18, 2021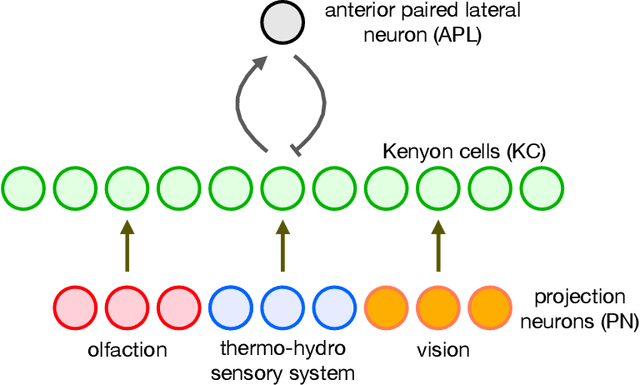
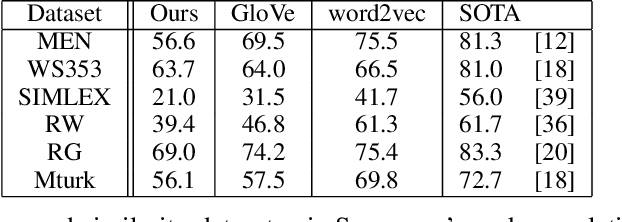
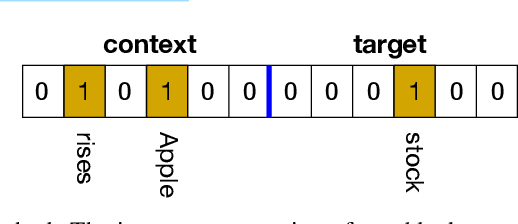
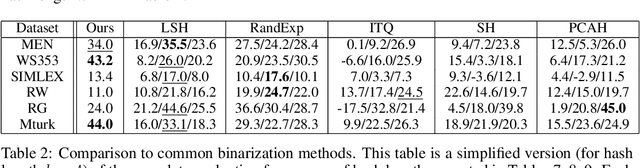
Abstract:The mushroom body of the fruit fly brain is one of the best studied systems in neuroscience. At its core it consists of a population of Kenyon cells, which receive inputs from multiple sensory modalities. These cells are inhibited by the anterior paired lateral neuron, thus creating a sparse high dimensional representation of the inputs. In this work we study a mathematical formalization of this network motif and apply it to learning the correlational structure between words and their context in a corpus of unstructured text, a common natural language processing (NLP) task. We show that this network can learn semantic representations of words and can generate both static and context-dependent word embeddings. Unlike conventional methods (e.g., BERT, GloVe) that use dense representations for word embedding, our algorithm encodes semantic meaning of words and their context in the form of sparse binary hash codes. The quality of the learned representations is evaluated on word similarity analysis, word-sense disambiguation, and document classification. It is shown that not only can the fruit fly network motif achieve performance comparable to existing methods in NLP, but, additionally, it uses only a fraction of the computational resources (shorter training time and smaller memory footprint).
Bio-Inspired Hashing for Unsupervised Similarity Search
Jan 14, 2020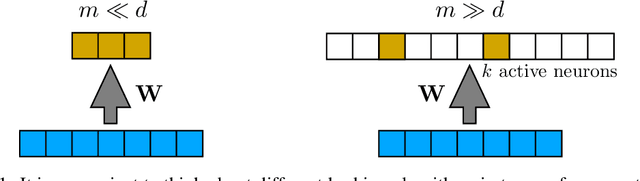
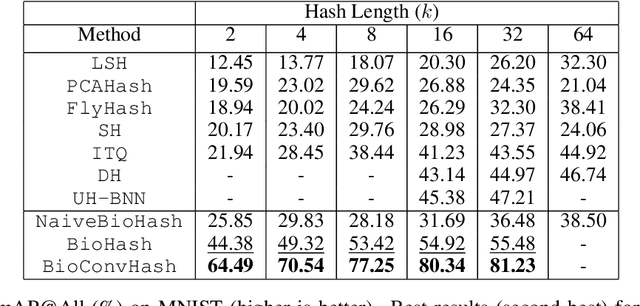
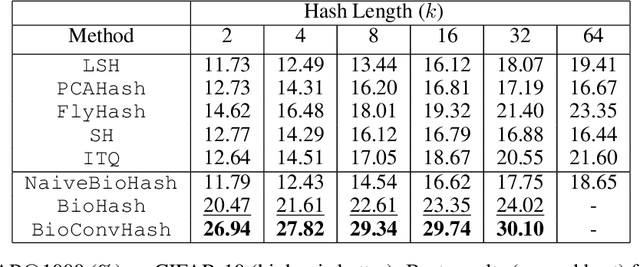
Abstract:The fruit fly Drosophila's olfactory circuit has inspired a new locality sensitive hashing (LSH) algorithm, FlyHash. In contrast with classical LSH algorithms that produce low dimensional hash codes, FlyHash produces sparse high-dimensional hash codes and has also been shown to have superior empirical performance compared to classical LSH algorithms in similarity search. However, FlyHash uses random projections and cannot learn from data. Building on inspiration from FlyHash and the ubiquity of sparse expansive representations in neurobiology, our work proposes a novel hashing algorithm BioHash that produces sparse high dimensional hash codes in a data-driven manner. We show that BioHash outperforms previously published benchmarks for various hashing methods. Since our learning algorithm is based on a local and biologically plausible synaptic plasticity rule, our work provides evidence for the proposal that LSH might be a computational reason for the abundance of sparse expansive motifs in a variety of biological systems. We also propose a convolutional variant BioConvHash that further improves performance. From the perspective of computer science, BioHash and BioConvHash are fast, scalable and yield compressed binary representations that are useful for similarity search.
Local Unsupervised Learning for Image Analysis
Aug 14, 2019
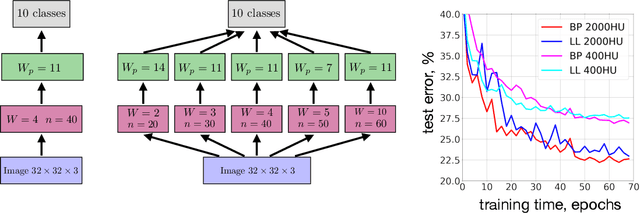
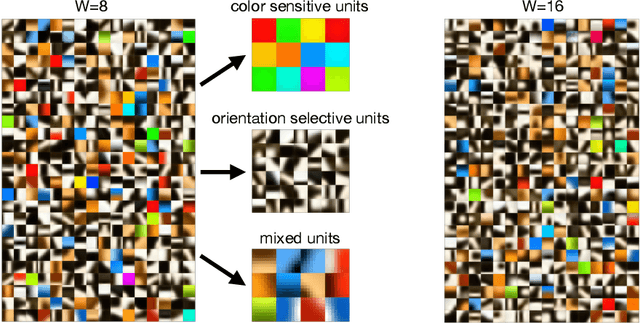
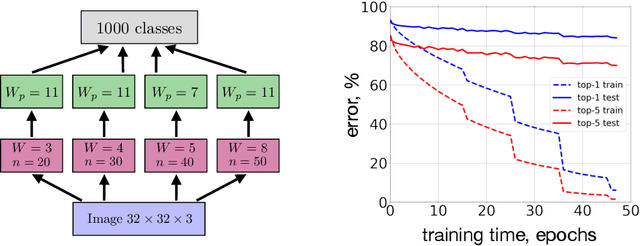
Abstract:Local Hebbian learning is believed to be inferior in performance to end-to-end training using a backpropagation algorithm. We question this popular belief by designing a local algorithm that can learn convolutional filters at scale on large image datasets. These filters combined with patch normalization and very steep non-linearities result in a good classification accuracy for shallow networks trained locally, as opposed to end-to-end. The filters learned by our algorithm contain both orientation selective units and unoriented color units, resembling the responses of pyramidal neurons located in the cytochrome oxidase 'interblob' and 'blob' regions in the primary visual cortex of primates. It is shown that convolutional networks with patch normalization significantly outperform standard convolutional networks on the task of recovering the original classes when shadows are superimposed on top of standard CIFAR-10 images. Patch normalization approximates the retinal adaptation to the mean light intensity, important for human vision. We also demonstrate a successful transfer of learned representations between CIFAR-10 and ImageNet 32x32 datasets. All these results taken together hint at the possibility that local unsupervised training might be a powerful tool for learning general representations (without specifying the task) directly from unlabeled data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge