Lei Jia
Reducing Memory Contention and I/O Congestion for Disk-based GNN Training
Jun 20, 2024Abstract:Graph neural networks (GNNs) gain wide popularity. Large graphs with high-dimensional features become common and training GNNs on them is non-trivial on an ordinary machine. Given a gigantic graph, even sample-based GNN training cannot work efficiently, since it is difficult to keep the graph's entire data in memory during the training process. Leveraging a solid-state drive (SSD) or other storage devices to extend the memory space has been studied in training GNNs. Memory and I/Os are hence critical for effectual disk-based training. We find that state-of-the-art (SoTA) disk-based GNN training systems severely suffer from issues like the memory contention between a graph's topological and feature data, and severe I/O congestion upon loading data from SSD for training. We accordingly develop GNNDrive. GNNDrive 1) minimizes the memory footprint with holistic buffer management across sampling and extracting, and 2) avoids I/O congestion through a strategy of asynchronous feature extraction. It also avoids costly data preparation on the critical path and makes the most of software and hardware resources. Experiments show that GNNDrive achieves superior performance. For example, when training with the Papers100M dataset and GraphSAGE model, GNNDrive is faster than SoTA PyG+, Ginex, and MariusGNN by 16.9x, 2.6x, and 2.7x, respectively.
Kernel-Induced Label Propagation by Mapping for Semi-Supervised Classification
May 31, 2019



Abstract:Kernel methods have been successfully applied to the areas of pattern recognition and data mining. In this paper, we mainly discuss the issue of propagating labels in kernel space. A Kernel-Induced Label Propagation (Kernel-LP) framework by mapping is proposed for high-dimensional data classification using the most informative patterns of data in kernel space. The essence of Kernel-LP is to perform joint label propagation and adaptive weight learning in a transformed kernel space. That is, our Kernel-LP changes the task of label propagation from the commonly-used Euclidean space in most existing work to kernel space. The motivation of our Kernel-LP to propagate labels and learn the adaptive weights jointly by the assumption of an inner product space of inputs, i.e., the original linearly inseparable inputs may be mapped to be separable in kernel space. Kernel-LP is based on existing positive and negative LP model, i.e., the effects of negative label information are integrated to improve the label prediction power. Also, Kernel-LP performs adaptive weight construction over the same kernel space, so it can avoid the tricky process of choosing the optimal neighborhood size suffered in traditional criteria. Two novel and efficient out-of-sample approaches for our Kernel-LP to involve new test data are also presented, i.e., (1) direct kernel mapping and (2) kernel mapping-induced label reconstruction, both of which purely depend on the kernel matrix between training set and testing set. Owing to the kernel trick, our algorithms will be applicable to handle the high-dimensional real data. Extensive results on real datasets demonstrate the effectiveness of our approach.
Chemi-net: a graph convolutional network for accurate drug property prediction
Mar 21, 2018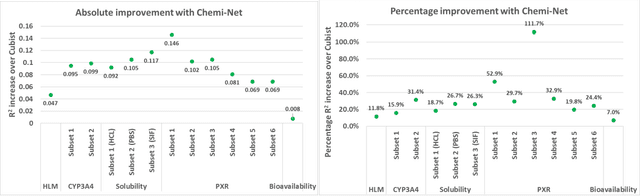
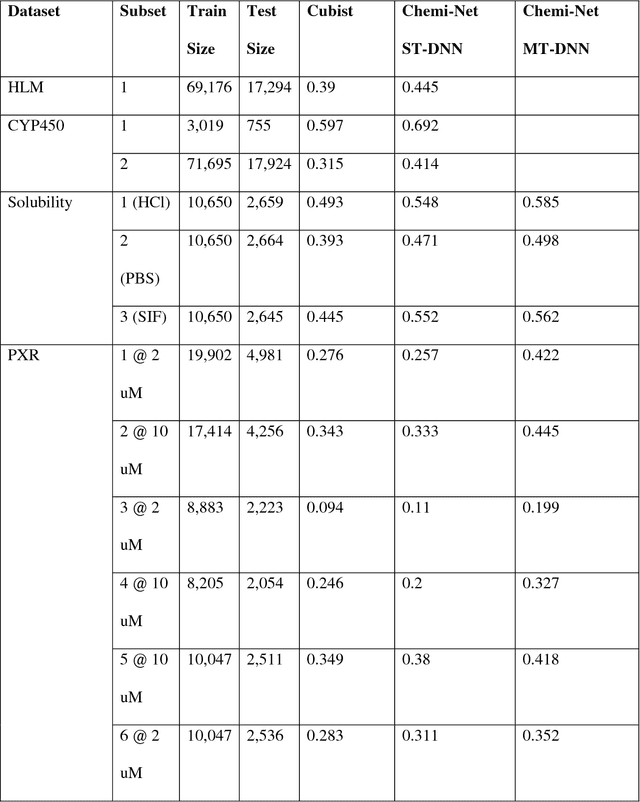
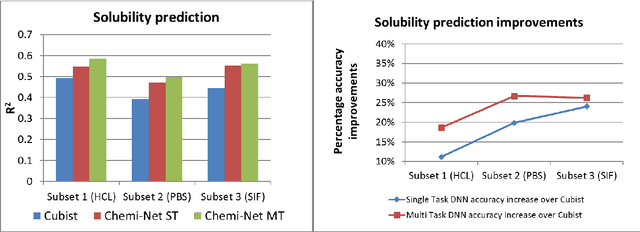
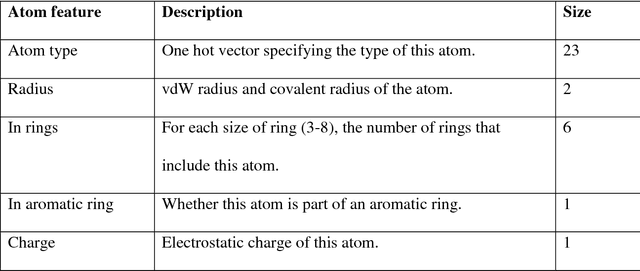
Abstract:Absorption, distribution, metabolism, and excretion (ADME) studies are critical for drug discovery. Conventionally, these tasks, together with other chemical property predictions, rely on domain-specific feature descriptors, or fingerprints. Following the recent success of neural networks, we developed Chemi-Net, a completely data-driven, domain knowledge-free, deep learning method for ADME property prediction. To compare the relative performance of Chemi-Net with Cubist, one of the popular machine learning programs used by Amgen, a large-scale ADME property prediction study was performed on-site at Amgen. The results showed that our deep neural network method improved current methods by a large margin. We foresee that the significantly increased accuracy of ADME prediction seen with Chemi-Net over Cubist will greatly accelerate drug discovery.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge