Kurt Stockinger
Institute of Computer Science, Zurich University of Applied Sciences, Winterthur, Switzerland
Query Carefully: Detecting the Unanswerables in Text-to-SQL Tasks
Dec 19, 2025Abstract:Text-to-SQL systems allow non-SQL experts to interact with relational databases using natural language. However, their tendency to generate executable SQL for ambiguous, out-of-scope, or unanswerable queries introduces a hidden risk, as outputs may be misinterpreted as correct. This risk is especially serious in biomedical contexts, where precision is critical. We therefore present Query Carefully, a pipeline that integrates LLM-based SQL generation with explicit detection and handling of unanswerable inputs. Building on the OncoMX component of ScienceBenchmark, we construct OncoMX-NAQ (No-Answer Questions), a set of 80 no-answer questions spanning 8 categories (non-SQL, out-of-schema/domain, and multiple ambiguity types). Our approach employs llama3.3:70b with schema-aware prompts, explicit No-Answer Rules (NAR), and few-shot examples drawn from both answerable and unanswerable questions. We evaluate SQL exact match, result accuracy, and unanswerable-detection accuracy. On the OncoMX dev split, few-shot prompting with answerable examples increases result accuracy, and adding unanswerable examples does not degrade performance. On OncoMX-NAQ, balanced prompting achieves the highest unanswerable-detection accuracy (0.8), with near-perfect results for structurally defined categories (non-SQL, missing columns, out-of-domain) but persistent challenges for missing-value queries (0.5) and column ambiguity (0.3). A lightweight user interface surfaces interim SQL, execution results, and abstentions, supporting transparent and reliable text-to-SQL in biomedical applications.
Quanvolutional Neural Networks for Spectrum Peak-Finding
Dec 15, 2025Abstract:The analysis of spectra, such as Nuclear Magnetic Resonance (NMR) spectra, for the comprehensive characterization of peaks is a challenging task for both experts and machines, especially with complex molecules. This process, also known as deconvolution, involves identifying and quantifying the peaks in the spectrum. Machine learning techniques have shown promising results in automating this process. With the advent of quantum computing, there is potential to further enhance these techniques. In this work, inspired by the success of classical Convolutional Neural Networks (CNNs), we explore the use of Quanvolutional Neural Networks (QuanvNNs) for the multi-task peak finding problem, involving both peak counting and position estimation. We implement a simple and interpretable QuanvNN architecture that can be directly compared to its classical CNN counterpart, and evaluate its performance on a synthetic NMR-inspired dataset. Our results demonstrate that QuanvNNs outperform classical CNNs on challenging spectra, achieving an 11\% improvement in F1 score and a 30\% reduction in mean absolute error for peak position estimation. Additionally, QuanvNNs appear to exhibit better convergence stability for harder problems.
TransClean: Finding False Positives in Multi-Source Entity Matching under Real-World Conditions via Transitive Consistency
Jun 04, 2025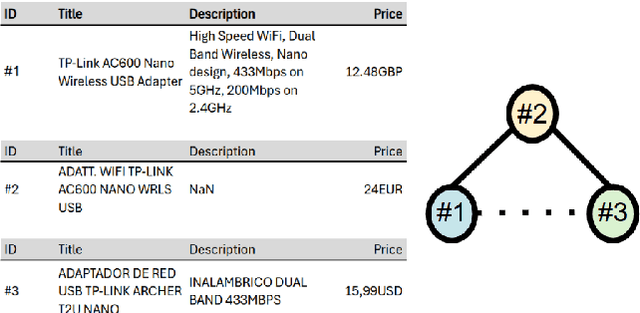
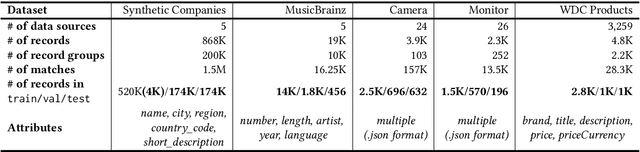
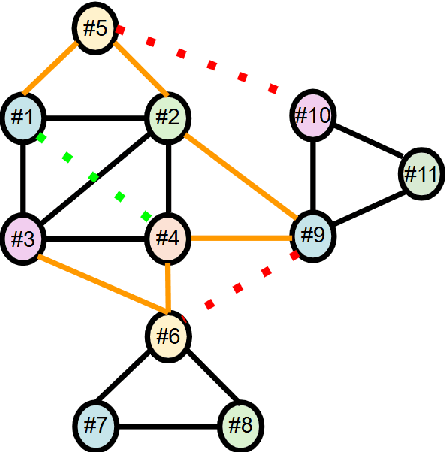
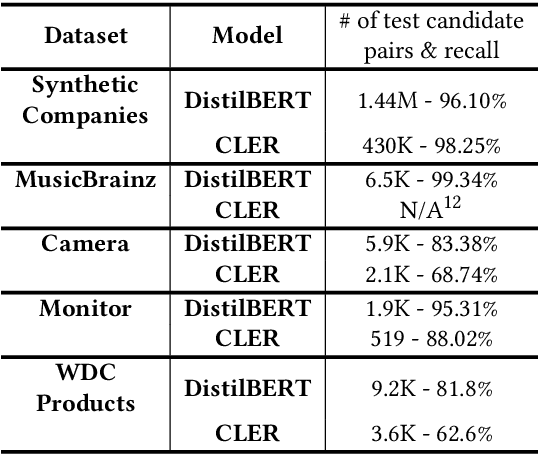
Abstract:We present TransClean, a method for detecting false positive predictions of entity matching algorithms under real-world conditions characterized by large-scale, noisy, and unlabeled multi-source datasets that undergo distributional shifts. TransClean is explicitly designed to operate with multiple data sources in an efficient, robust and fast manner while accounting for edge cases and requiring limited manual labeling. TransClean leverages the Transitive Consistency of a matching, a measure of the consistency of a pairwise matching model f_theta on the matching it produces G_f_theta, based both on its predictions on directly evaluated record pairs and its predictions on implied record pairs. TransClean iteratively modifies a matching through gradually removing false positive matches while removing as few true positive matches as possible. In each of these steps, the estimation of the Transitive Consistency is exclusively done through model evaluations and produces quantities that can be used as proxies of the amounts of true and false positives in the matching while not requiring any manual labeling, producing an estimate of the quality of the matching and indicating which record groups are likely to contain false positives. In our experiments, we compare combining TransClean with a naively trained pairwise matching model (DistilBERT) and with a state-of-the-art end-to-end matching method (CLER) and illustrate the flexibility of TransClean in being able to detect most of the false positives of either setup across a variety of datasets. Our experiments show that TransClean induces an average +24.42 F1 score improvement for entity matching in a multi-source setting when compared to traditional pair-wise matching algorithms.
Explainable Multi-Modal Data Exploration in Natural Language via LLM Agent
Dec 24, 2024



Abstract:International enterprises, organizations, or hospitals collect large amounts of multi-modal data stored in databases, text documents, images, and videos. While there has been recent progress in the separate fields of multi-modal data exploration as well as in database systems that automatically translate natural language questions to database query languages, the research challenge of querying database systems combined with other unstructured modalities such as images in natural language is widely unexplored. In this paper, we propose XMODE - a system that enables explainable, multi-modal data exploration in natural language. Our approach is based on the following research contributions: (1) Our system is inspired by a real-world use case that enables users to explore multi-modal information systems. (2) XMODE leverages a LLM-based agentic AI framework to decompose a natural language question into subtasks such as text-to-SQL generation and image analysis. (3) Experimental results on multi-modal datasets over relational data and images demonstrate that our system outperforms state-of-the-art multi-modal exploration systems, excelling not only in accuracy but also in various performance metrics such as query latency, API costs, planning efficiency, and explanation quality, thanks to the more effective utilization of the reasoning capabilities of LLMs.
SM3-Text-to-Query: Synthetic Multi-Model Medical Text-to-Query Benchmark
Nov 08, 2024


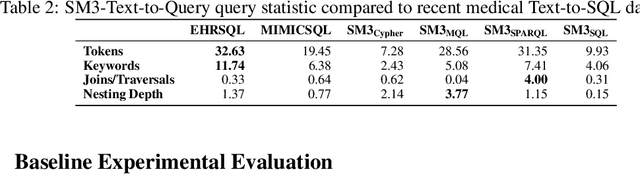
Abstract:Electronic health records (EHRs) are stored in various database systems with different database models on heterogeneous storage architectures, such as relational databases, document stores, or graph databases. These different database models have a big impact on query complexity and performance. While this has been a known fact in database research, its implications for the growing number of Text-to-Query systems have surprisingly not been investigated so far. In this paper, we present SM3-Text-to-Query, the first multi-model medical Text-to-Query benchmark based on synthetic patient data from Synthea, following the SNOMED-CT taxonomy -- a widely used knowledge graph ontology covering medical terminology. SM3-Text-to-Query provides data representations for relational databases (PostgreSQL), document stores (MongoDB), and graph databases (Neo4j and GraphDB (RDF)), allowing the evaluation across four popular query languages, namely SQL, MQL, Cypher, and SPARQL. We systematically and manually develop 408 template questions, which we augment to construct a benchmark of 10K diverse natural language question/query pairs for these four query languages (40K pairs overall). On our dataset, we evaluate several common in-context-learning (ICL) approaches for a set of representative closed and open-source LLMs. Our evaluation sheds light on the trade-offs between database models and query languages for different ICL strategies and LLMs. Last, SM3-Text-to-Query is easily extendable to additional query languages or real, standard-based patient databases.
GenJoin: Conditional Generative Plan-to-Plan Query Optimizer that Learns from Subplan Hints
Nov 07, 2024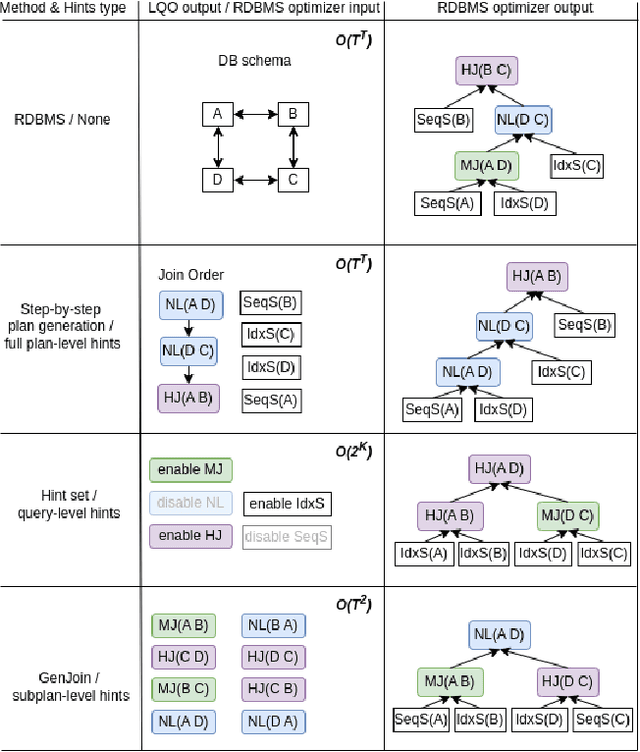
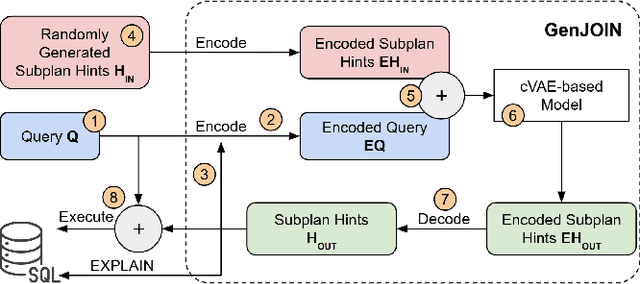
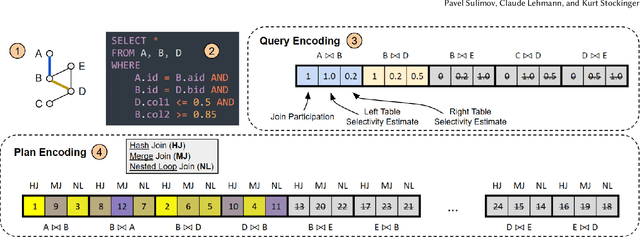
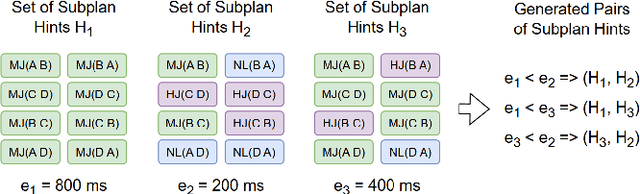
Abstract:Query optimization has become a research area where classical algorithms are being challenged by machine learning algorithms. At the same time, recent trends in learned query optimizers have shown that it is prudent to take advantage of decades of database research and augment classical query optimizers by shrinking the plan search space through different types of hints (e.g. by specifying the join type, scan type or the order of joins) rather than completely replacing the classical query optimizer with machine learning models. It is especially relevant for cases when classical optimizers cannot fully enumerate all logical and physical plans and, as an alternative, need to rely on less robust approaches like genetic algorithms. However, even symbiotically learned query optimizers are hampered by the need for vast amounts of training data, slow plan generation during inference and unstable results across various workload conditions. In this paper, we present GenJoin - a novel learned query optimizer that considers the query optimization problem as a generative task and is capable of learning from a random set of subplan hints to produce query plans that outperform the classical optimizer. GenJoin is the first learned query optimizer that significantly and consistently outperforms PostgreSQL as well as state-of-the-art methods on two well-known real-world benchmarks across a variety of workloads using rigorous machine learning evaluations.
Applying Quantum Autoencoders for Time Series Anomaly Detection
Oct 05, 2024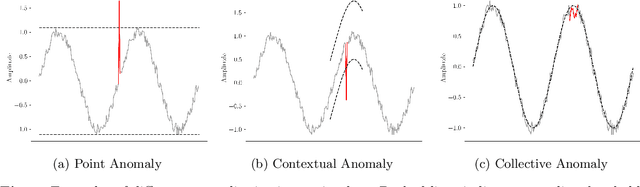
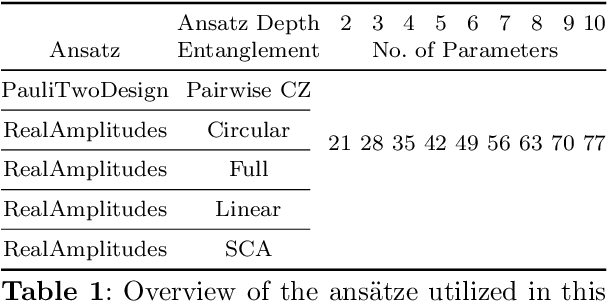
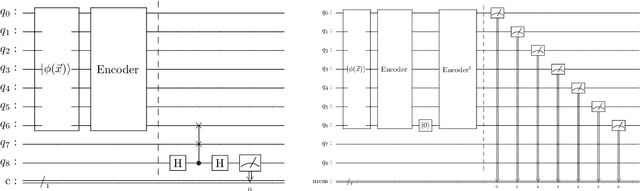
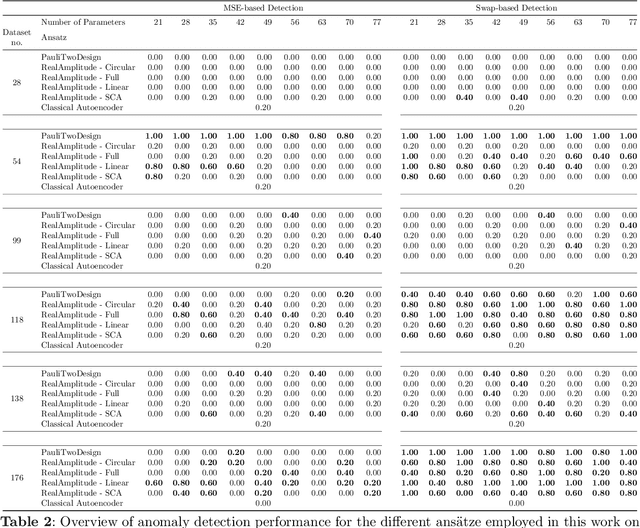
Abstract:Anomaly detection is an important problem with applications in various domains such as fraud detection, pattern recognition or medical diagnosis. Several algorithms have been introduced using classical computing approaches. However, using quantum computing for solving anomaly detection problems in time series data is a widely unexplored research field. This paper explores the application of quantum autoencoders to time series anomaly detection. We investigate two primary techniques for classifying anomalies: (1) Analyzing the reconstruction error generated by the quantum autoencoder and (2) latent representation analysis. Our simulated experimental results, conducted across various ansaetze, demonstrate that quantum autoencoders consistently outperform classical deep learning-based autoencoders across multiple datasets. Specifically, quantum autoencoders achieve superior anomaly detection performance while utilizing 60-230 times fewer parameters and requiring five times fewer training iterations. In addition, we implement our quantum encoder on real quantum hardware. Our experimental results demonstrate that quantum autoencoders achieve anomaly detection performance on par with their simulated counterparts.
GraLMatch: Matching Groups of Entities with Graphs and Language Models
Jun 21, 2024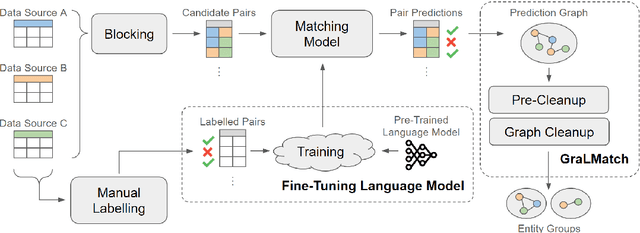
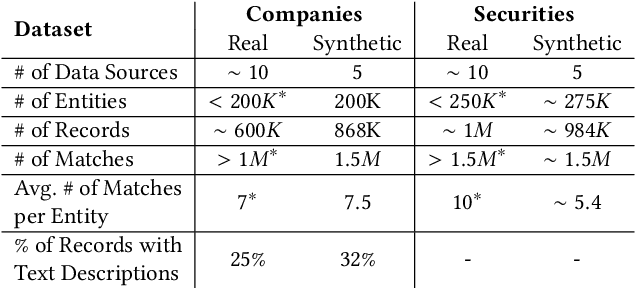
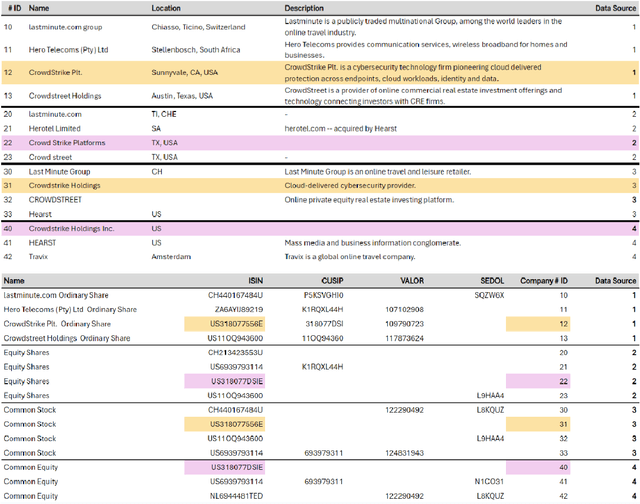
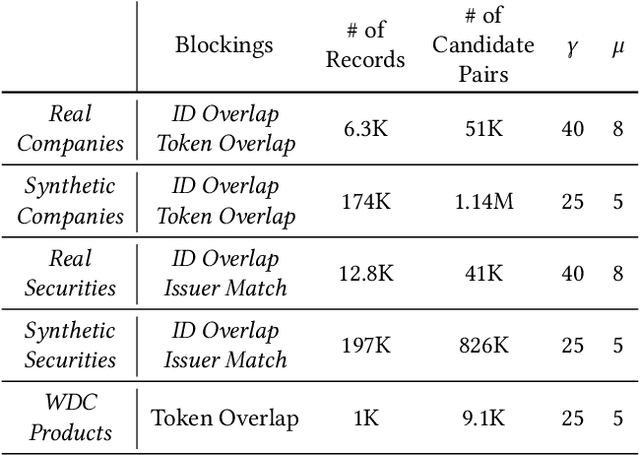
Abstract:In this paper, we present an end-to-end multi-source Entity Matching problem, which we call entity group matching, where the goal is to assign to the same group, records originating from multiple data sources but representing the same real-world entity. We focus on the effects of transitively matched records, i.e. the records connected by paths in the graph G = (V,E) whose nodes and edges represent the records and whether they are a match or not. We present a real-world instance of this problem, where the challenge is to match records of companies and financial securities originating from different data providers. We also introduce two new multi-source benchmark datasets that present similar matching challenges as real-world records. A distinctive characteristic of these records is that they are regularly updated following real-world events, but updates are not applied uniformly across data sources. This phenomenon makes the matching of certain groups of records only possible through the use of transitive information. In our experiments, we illustrate how considering transitively matched records is challenging since a limited amount of false positive pairwise match predictions can throw off the group assignment of large quantities of records. Thus, we propose GraLMatch, a method that can partially detect and remove false positive pairwise predictions through graph-based properties. Finally, we showcase how fine-tuning a Transformer-based model (DistilBERT) on a reduced number of labeled samples yields a better final entity group matching than training on more samples and/or incorporating fine-tuning optimizations, illustrating how precision becomes the deciding factor in the entity group matching of large volumes of records.
StatBot.Swiss: Bilingual Open Data Exploration in Natural Language
Jun 06, 2024



Abstract:The potential for improvements brought by Large Language Models (LLMs) in Text-to-SQL systems is mostly assessed on monolingual English datasets. However, LLMs' performance for other languages remains vastly unexplored. In this work, we release the StatBot.Swiss dataset, the first bilingual benchmark for evaluating Text-to-SQL systems based on real-world applications. The StatBot.Swiss dataset contains 455 natural language/SQL-pairs over 35 big databases with varying level of complexity for both English and German. We evaluate the performance of state-of-the-art LLMs such as GPT-3.5-Turbo and mixtral-8x7b-instruct for the Text-to-SQL translation task using an in-context learning approach. Our experimental analysis illustrates that current LLMs struggle to generalize well in generating SQL queries on our novel bilingual dataset.
Evaluating the Data Model Robustness of Text-to-SQL Systems Based on Real User Queries
Feb 13, 2024



Abstract:Text-to-SQL systems (also known as NL-to-SQL systems) have become an increasingly popular solution for bridging the gap between user capabilities and SQL-based data access. These systems translate user requests in natural language to valid SQL statements for a specific database. Recent Text-to-SQL systems have benefited from the rapid improvement of transformer-based language models. However, while Text-to-SQL systems that incorporate such models continuously reach new high scores on -- often synthetic -- benchmark datasets, a systematic exploration of their robustness towards different data models in a real-world, realistic scenario is notably missing. This paper provides the first in-depth evaluation of the data model robustness of Text-to-SQL systems in practice based on a multi-year international project focused on Text-to-SQL interfaces. Our evaluation is based on a real-world deployment of FootballDB, a system that was deployed over a 9 month period in the context of the FIFA World Cup 2022, during which about 6K natural language questions were asked and executed. All of our data is based on real user questions that were asked live to the system. We manually labeled and translated a subset of these questions for three different data models. For each data model, we explore the performance of representative Text-to-SQL systems and language models. We further quantify the impact of training data size, pre-, and post-processing steps as well as language model inference time. Our comprehensive evaluation sheds light on the design choices of real-world Text-to-SQL systems and their impact on moving from research prototypes to real deployments. Last, we provide a new benchmark dataset to the community, which is the first to enable the evaluation of different data models for the same dataset and is substantially more challenging than most previous datasets in terms of query complexity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge