Ellery Smith
ZHAW Zurich University of Applied Sciences, Switzerland
StatBot.Swiss: Bilingual Open Data Exploration in Natural Language
Jun 06, 2024



Abstract:The potential for improvements brought by Large Language Models (LLMs) in Text-to-SQL systems is mostly assessed on monolingual English datasets. However, LLMs' performance for other languages remains vastly unexplored. In this work, we release the StatBot.Swiss dataset, the first bilingual benchmark for evaluating Text-to-SQL systems based on real-world applications. The StatBot.Swiss dataset contains 455 natural language/SQL-pairs over 35 big databases with varying level of complexity for both English and German. We evaluate the performance of state-of-the-art LLMs such as GPT-3.5-Turbo and mixtral-8x7b-instruct for the Text-to-SQL translation task using an in-context learning approach. Our experimental analysis illustrates that current LLMs struggle to generalize well in generating SQL queries on our novel bilingual dataset.
Data-Driven Information Extraction and Enrichment of Molecular Profiling Data for Cancer Cell Lines
Jul 03, 2023Abstract:With the proliferation of research means and computational methodologies, published biomedical literature is growing exponentially in numbers and volume. As a consequence, in the fields of biological, medical and clinical research, domain experts have to sift through massive amounts of scientific text to find relevant information. However, this process is extremely tedious and slow to be performed by humans. Hence, novel computational information extraction and correlation mechanisms are required to boost meaningful knowledge extraction. In this work, we present the design, implementation and application of a novel data extraction and exploration system. This system extracts deep semantic relations between textual entities from scientific literature to enrich existing structured clinical data in the domain of cancer cell lines. We introduce a new public data exploration portal, which enables automatic linking of genomic copy number variants plots with ranked, related entities such as affected genes. Each relation is accompanied by literature-derived evidences, allowing for deep, yet rapid, literature search, using existing structured data as a springboard. Our system is publicly available on the web at https://cancercelllines.org
INODE: Building an End-to-End Data Exploration System in Practice
Apr 09, 2021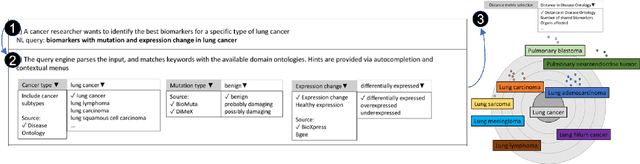
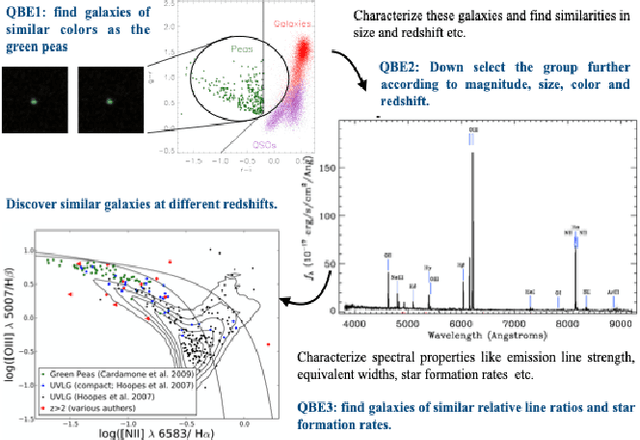
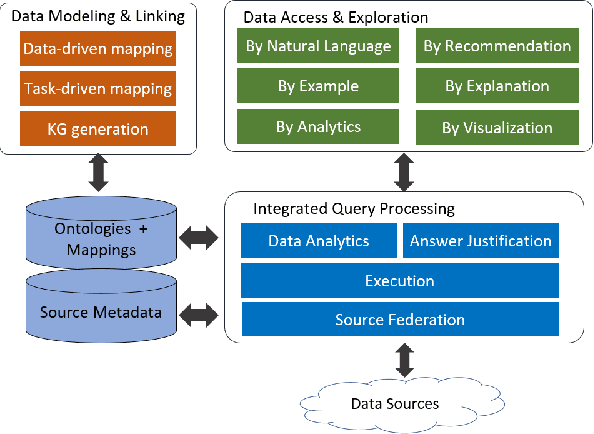
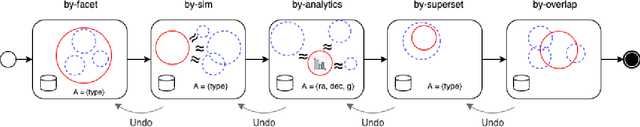
Abstract:A full-fledged data exploration system must combine different access modalities with a powerful concept of guiding the user in the exploration process, by being reactive and anticipative both for data discovery and for data linking. Such systems are a real opportunity for our community to cater to users with different domain and data science expertise. We introduce INODE -- an end-to-end data exploration system -- that leverages, on the one hand, Machine Learning and, on the other hand, semantics for the purpose of Data Management (DM). Our vision is to develop a classic unified, comprehensive platform that provides extensive access to open datasets, and we demonstrate it in three significant use cases in the fields of Cancer Biomarker Reearch, Research and Innovation Policy Making, and Astrophysics. INODE offers sustainable services in (a) data modeling and linking, (b) integrated query processing using natural language, (c) guidance, and (d) data exploration through visualization, thus facilitating the user in discovering new insights. We demonstrate that our system is uniquely accessible to a wide range of users from larger scientific communities to the public. Finally, we briefly illustrate how this work paves the way for new research opportunities in DM.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge