Sithursan Sivasubramaniam
SM3-Text-to-Query: Synthetic Multi-Model Medical Text-to-Query Benchmark
Nov 08, 2024


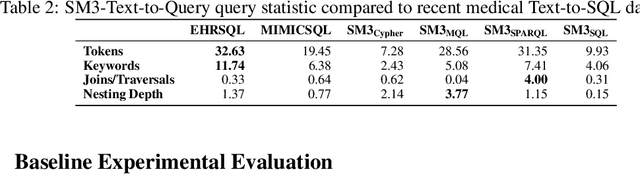
Abstract:Electronic health records (EHRs) are stored in various database systems with different database models on heterogeneous storage architectures, such as relational databases, document stores, or graph databases. These different database models have a big impact on query complexity and performance. While this has been a known fact in database research, its implications for the growing number of Text-to-Query systems have surprisingly not been investigated so far. In this paper, we present SM3-Text-to-Query, the first multi-model medical Text-to-Query benchmark based on synthetic patient data from Synthea, following the SNOMED-CT taxonomy -- a widely used knowledge graph ontology covering medical terminology. SM3-Text-to-Query provides data representations for relational databases (PostgreSQL), document stores (MongoDB), and graph databases (Neo4j and GraphDB (RDF)), allowing the evaluation across four popular query languages, namely SQL, MQL, Cypher, and SPARQL. We systematically and manually develop 408 template questions, which we augment to construct a benchmark of 10K diverse natural language question/query pairs for these four query languages (40K pairs overall). On our dataset, we evaluate several common in-context-learning (ICL) approaches for a set of representative closed and open-source LLMs. Our evaluation sheds light on the trade-offs between database models and query languages for different ICL strategies and LLMs. Last, SM3-Text-to-Query is easily extendable to additional query languages or real, standard-based patient databases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge