Kerui Zhu
Investigating Instruction Tuning Large Language Models on Graphs
Aug 10, 2024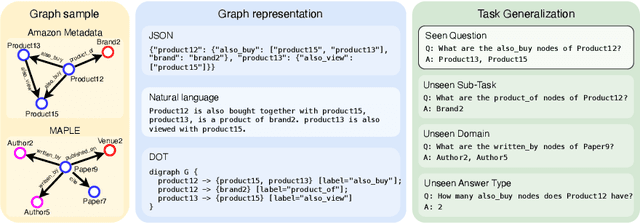
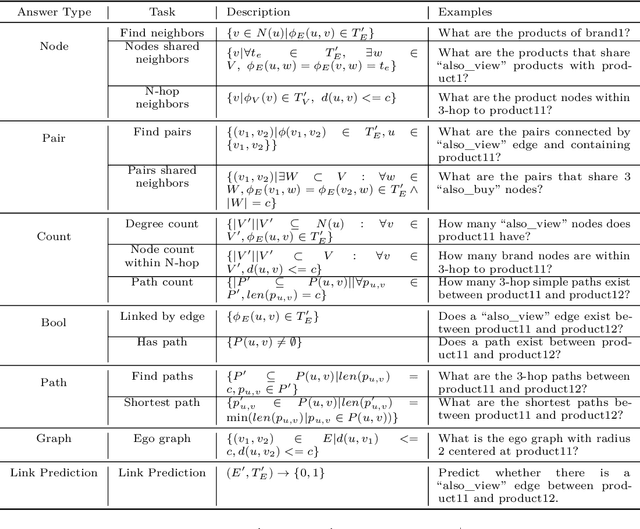

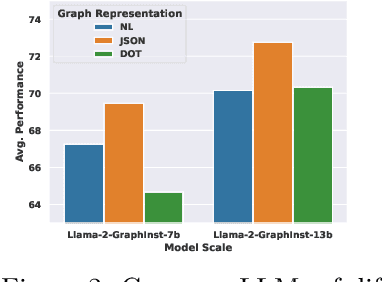
Abstract:Inspired by the recent advancements of Large Language Models (LLMs) in NLP tasks, there's growing interest in applying LLMs to graph-related tasks. This study delves into the capabilities of instruction-following LLMs for engaging with real-world graphs, aiming to offer empirical insights into how LLMs can effectively interact with graphs and generalize across graph tasks. We begin by constructing a dataset designed for instruction tuning, which comprises a diverse collection of 79 graph-related tasks from academic and e-commerce domains, featuring 44,240 training instances and 18,960 test samples. Utilizing this benchmark, our initial investigation focuses on identifying the optimal graph representation that serves as a conduit for LLMs to understand complex graph structures. Our findings indicate that JSON format for graph representation consistently outperforms natural language and code formats across various LLMs and graph types. Furthermore, we examine the key factors that influence the generalization abilities of instruction-tuned LLMs by evaluating their performance on both in-domain and out-of-domain graph tasks.
Descriptive Knowledge Graph in Biomedical Domain
Oct 18, 2023



Abstract:We present a novel system that automatically extracts and generates informative and descriptive sentences from the biomedical corpus and facilitates the efficient search for relational knowledge. Unlike previous search engines or exploration systems that retrieve unconnected passages, our system organizes descriptive sentences as a relational graph, enabling researchers to explore closely related biomedical entities (e.g., diseases treated by a chemical) or indirectly connected entities (e.g., potential drugs for treating a disease). Our system also uses ChatGPT and a fine-tuned relation synthesis model to generate concise and reliable descriptive sentences from retrieved information, reducing the need for extensive human reading effort. With our system, researchers can easily obtain both high-level knowledge and detailed references and interactively steer to the information of interest. We spotlight the application of our system in COVID-19 research, illustrating its utility in areas such as drug repurposing and literature curation.
DimonGen: Diversified Generative Commonsense Reasoning for Explaining Concept Relationships
Dec 20, 2022



Abstract:In this paper, we propose DimonGen, which aims to generate diverse sentences describing concept relationships in various everyday scenarios. To support this, we create a benchmark dataset for this task by adapting the existing CommonGen dataset and propose a two-stage model called MoREE (Mixture of Retrieval-Enhanced Experts) to generate the target sentences. MoREE consists of a mixture of retriever models that retrieve diverse context sentences related to the given concepts, and a mixture of generator models that generate diverse sentences based on the retrieved contexts. We conduct experiments on the DimonGen task and show that MoREE outperforms strong baselines in terms of both the quality and diversity of the generated sentences. Our results demonstrate that MoREE is able to generate diverse sentences that reflect different relationships between concepts, leading to a comprehensive understanding of concept relationships.
DKG: A Descriptive Knowledge Graph for Explaining Relationships between Entities
May 21, 2022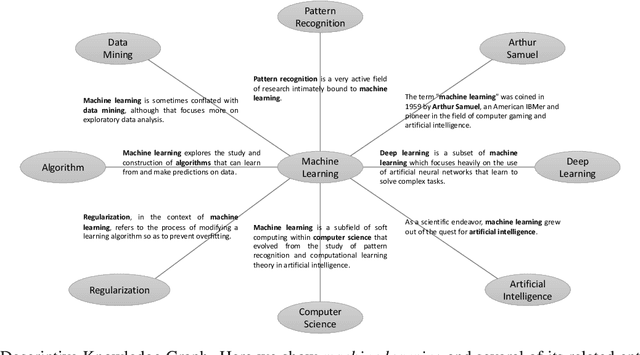
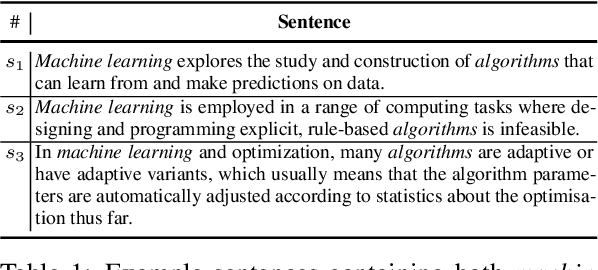
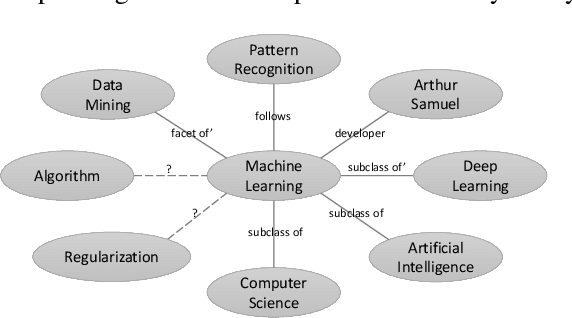
Abstract:In this paper, we propose Descriptive Knowledge Graph (DKG) - an open and interpretable form of modeling relationships between entities. In DKGs, relationships between entities are represented by relation descriptions. For instance, the relationship between entities of machine learning and algorithm can be described as "Machine learning explores the study and construction of algorithms that can learn from and make predictions on data." To construct DKGs, we propose a self-supervised learning method to extract relation descriptions with the analysis of dependency patterns and a transformer-based relation description synthesizing model to generate relation descriptions. Experiments demonstrate that our system can extract and generate high-quality relation descriptions for explaining entity relationships.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge