Kerstin Bunte
Bernoulli Institute of Mathematics, Computer Science and Artificial Intelligence, University of Groningen, The Netherlands
On the importance of structural identifiability for machine learning with partially observed dynamical systems
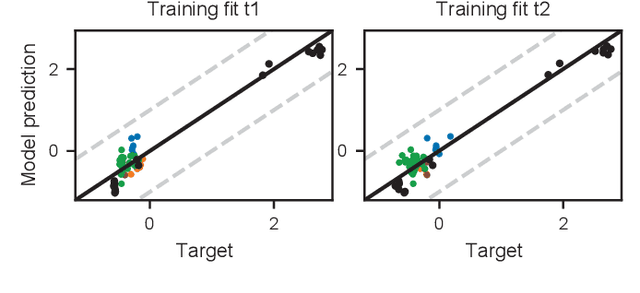
Feb 06, 2025Abstract:The successful application of modern machine learning for time series classification is often hampered by limitations in quality and quantity of available training data. To overcome these limitations, available domain expert knowledge in the form of parametrised mechanistic dynamical models can be used whenever it is available and time series observations may be represented as an element from a given class of parametrised dynamical models. This makes the learning process interpretable and allows the modeller to deal with sparsely and irregularly sampled data in a natural way. However, the internal processes of a dynamical model are often only partially observed. This can lead to ambiguity regarding which particular model realization best explains a given time series observation. This problem is well-known in the literature, and a dynamical model with this issue is referred to as structurally unidentifiable. Training a classifier that incorporates knowledge about a structurally unidentifiable dynamical model can negatively influence classification performance. To address this issue, we employ structural identifiability analysis to explicitly relate parameter configurations that are associated with identical system outputs. Using the derived relations in classifier training, we demonstrate that this method significantly improves the classifier's ability to generalize to unseen data on a number of example models from the biomedical domain. This effect is especially pronounced when the number of training instances is limited. Our results demonstrate the importance of accounting for structural identifiability, a topic that has received relatively little attention from the machine learning community.
An Industry 4.0 example: real-time quality control for steel-based mass production using Machine Learning on non-invasive sensor data
Jun 12, 2022


Abstract:Insufficient steel quality in mass production can cause extremely costly damage to tooling, production downtimes and low quality products. Automatic, fast and cheap strategies to estimate essential material properties for quality control, risk mitigation and the prediction of faults are highly desirable. In this work we analyse a high throughput production line of steel-based products. Currently, the material quality is checked using manual destructive testing, which is slow, wasteful and covers only a tiny fraction of the material. To achieve complete testing coverage our industrial collaborator developed a contactless, non-invasive, electromagnetic sensor to measure all material during production in real-time. Our contribution is three-fold: 1) We show in a controlled experiment that the sensor can distinguish steel with deliberately altered properties. 2) 48 steel coils were fully measured non-invasively and additional destructive tests were conducted on samples to serve as ground truth. A linear model is fitted to predict from the non-invasive measurements two key material properties (yield strength and tensile strength) that normally are obtained by destructive tests. The performance is evaluated in leave-one-coil-out cross-validation. 3) The resulting model is used to analyse the material properties and the relationship with logged product faults on real production data of ~108 km of processed material measured with the non-invasive sensor. The model achieves an excellent performance (F3-score of 0.95) predicting material running out of specifications for the tensile strength. The combination of model predictions and logged product faults shows that if a significant percentage of estimated yield stress values is out of specification, the risk of product faults is high. Our analysis demonstrates promising directions for real-time quality control, risk monitoring and fault detection.
Interpretable Models Capable of Handling Systematic Missingness in Imbalanced Classes and Heterogeneous Datasets
Jun 04, 2022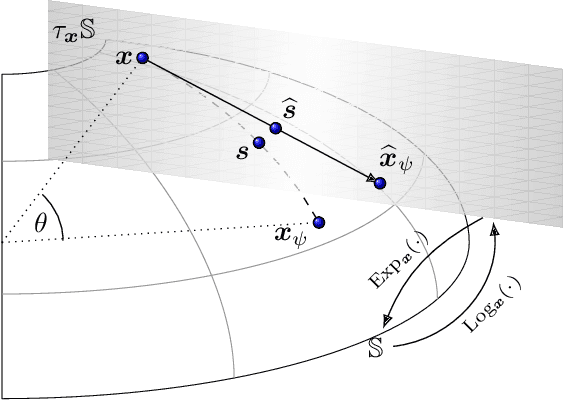
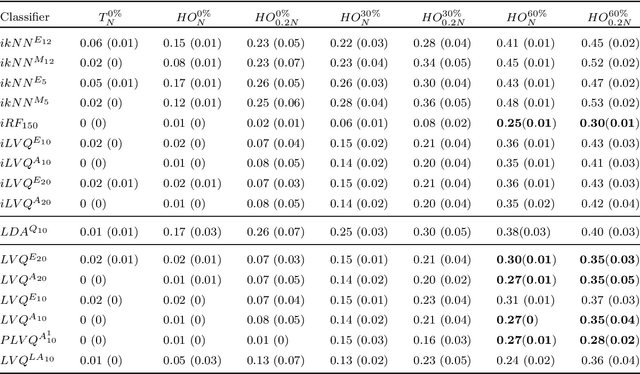

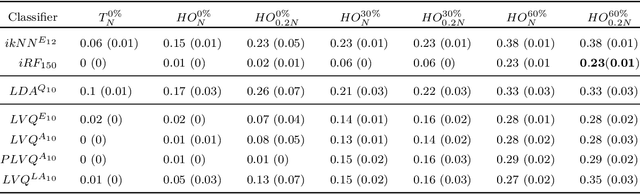
Abstract:Application of interpretable machine learning techniques on medical datasets facilitate early and fast diagnoses, along with getting deeper insight into the data. Furthermore, the transparency of these models increase trust among application domain experts. Medical datasets face common issues such as heterogeneous measurements, imbalanced classes with limited sample size, and missing data, which hinder the straightforward application of machine learning techniques. In this paper we present a family of prototype-based (PB) interpretable models which are capable of handling these issues. The models introduced in this contribution show comparable or superior performance to alternative techniques applicable in such situations. However, unlike ensemble based models, which have to compromise on easy interpretation, the PB models here do not. Moreover we propose a strategy of harnessing the power of ensembles while maintaining the intrinsic interpretability of the PB models, by averaging the model parameter manifolds. All the models were evaluated on a synthetic (publicly available dataset) in addition to detailed analyses of two real-world medical datasets (one publicly available). Results indicated that the models and strategies we introduced addressed the challenges of real-world medical data, while remaining computationally inexpensive and transparent, as well as similar or superior in performance compared to their alternatives.
Detection of extragalactic Ultra-Compact Dwarfs and Globular Clusters using Explainable AI techniques
Jan 07, 2022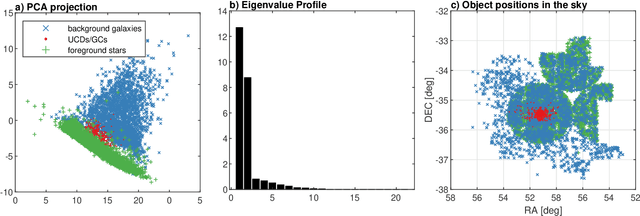
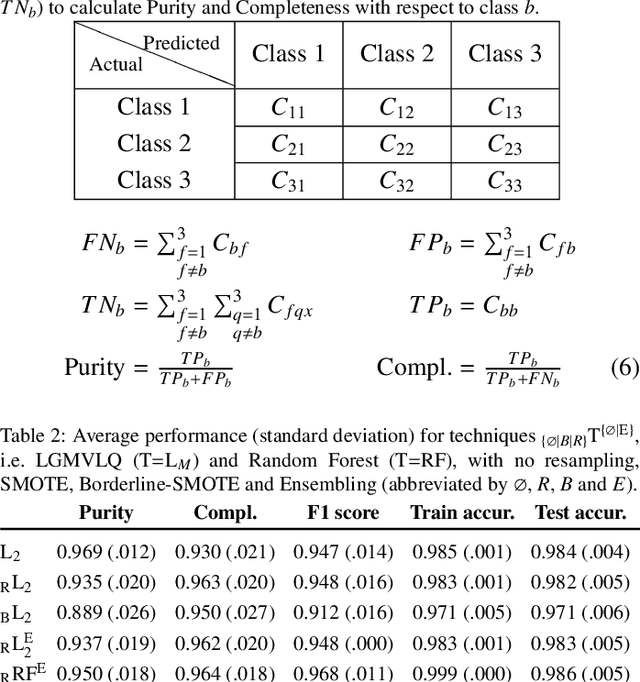
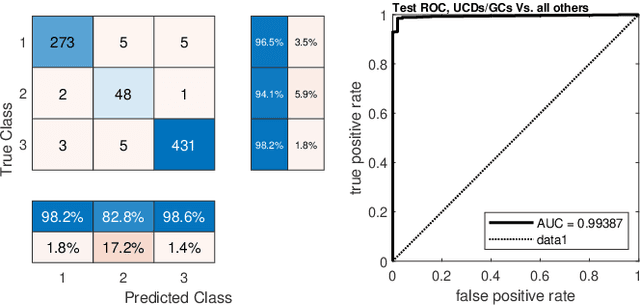
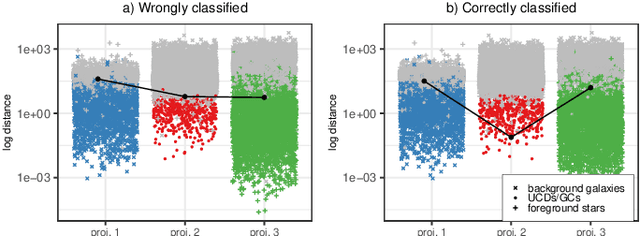
Abstract:Compact stellar systems such as Ultra-compact dwarfs (UCDs) and Globular Clusters (GCs) around galaxies are known to be the tracers of the merger events that have been forming these galaxies. Therefore, identifying such systems allows to study galaxies mass assembly, formation and evolution. However, in the lack of spectroscopic information detecting UCDs/GCs using imaging data is very uncertain. Here, we aim to train a machine learning model to separate these objects from the foreground stars and background galaxies using the multi-wavelength imaging data of the Fornax galaxy cluster in 6 filters, namely u, g, r, i, J and Ks. The classes of objects are highly imbalanced which is problematic for many automatic classification techniques. Hence, we employ Synthetic Minority Over-sampling to handle the imbalance of the training data. Then, we compare two classifiers, namely Localized Generalized Matrix Learning Vector Quantization (LGMLVQ) and Random Forest (RF). Both methods are able to identify UCDs/GCs with a precision and a recall of >93 percent and provide relevances that reflect the importance of each feature dimension %(colors and angular sizes) for the classification. Both methods detect angular sizes as important markers for this classification problem. While it is astronomical expectation that color indices of u-i and i-Ks are the most important colors, our analysis shows that colors such as g-r are more informative, potentially because of higher signal-to-noise ratio. Besides the excellent performance the LGMLVQ method allows further interpretability by providing the feature importance for each individual class, class-wise representative samples and the possibility for non-linear visualization of the data as demonstrated in this contribution. We conclude that employing machine learning techniques to identify UCDs/GCs can lead to promising results.
LAAT: Locally Aligned Ant Technique for detecting manifolds of varying density
Sep 17, 2020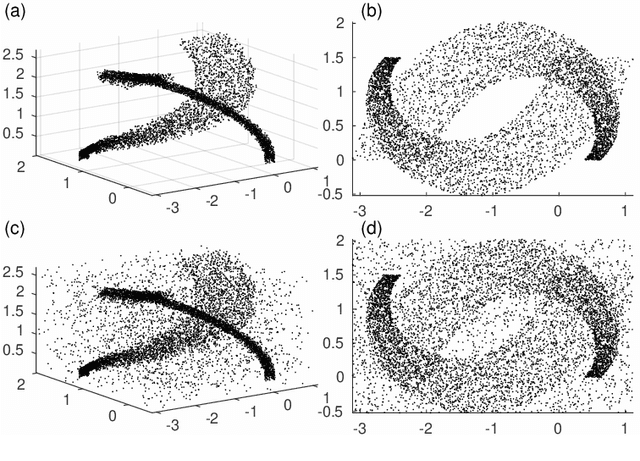


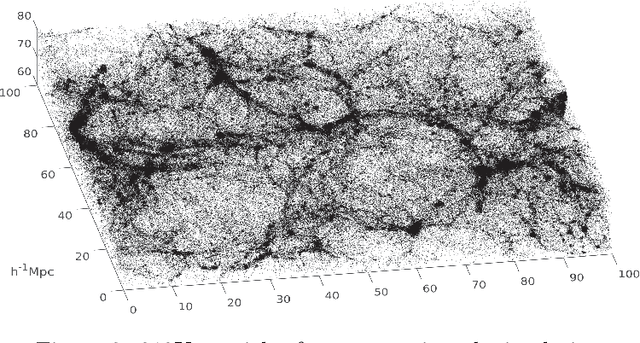
Abstract:Dimensionality reduction and clustering are often used as preliminary steps for many complex machine learning tasks. The presence of noise and outliers can deteriorate the performance of such preprocessing and therefore impair the subsequent analysis tremendously. In manifold learning, several studies indicate solutions for removing background noise or noise close to the structure when the density is substantially higher than that exhibited by the noise. However, in many applications, including astronomical datasets, the density varies alongside manifolds that are buried in a noisy background. We propose a novel method to extract manifolds in the presence of noise based on the idea of Ant colony optimization. In contrast to the existing random walk solutions, our technique captures points which are locally aligned with major directions of the manifold. Moreover, we empirically show that the biologically inspired formulation of ant pheromone reinforces this behavior enabling it to recover multiple manifolds embedded in extremely noisy data clouds. The algorithm's performance is demonstrated in comparison to the state-of-the-art approaches, such as Markov Chain, LLPD, and Disperse, on several synthetic and real astronomical datasets stemming from an N-body simulation of a cosmological volume.
Visualisation and knowledge discovery from interpretable models
May 08, 2020



Abstract:Increasing number of sectors which affect human lives, are using Machine Learning (ML) tools. Hence the need for understanding their working mechanism and evaluating their fairness in decision-making, are becoming paramount, ushering in the era of Explainable AI (XAI). In this contribution we introduced a few intrinsically interpretable models which are also capable of dealing with missing values, in addition to extracting knowledge from the dataset and about the problem. These models are also capable of visualisation of the classifier and decision boundaries: they are the angle based variants of Learning Vector Quantization. We have demonstrated the algorithms on a synthetic dataset and a real-world one (heart disease dataset from the UCI repository). The newly developed classifiers helped in investigating the complexities of the UCI dataset as a multiclass problem. The performance of the developed classifiers were comparable to those reported in literature for this dataset, with additional value of interpretability, when the dataset was treated as a binary class problem.
Sparse group factor analysis for biclustering of multiple data sources
Apr 21, 2016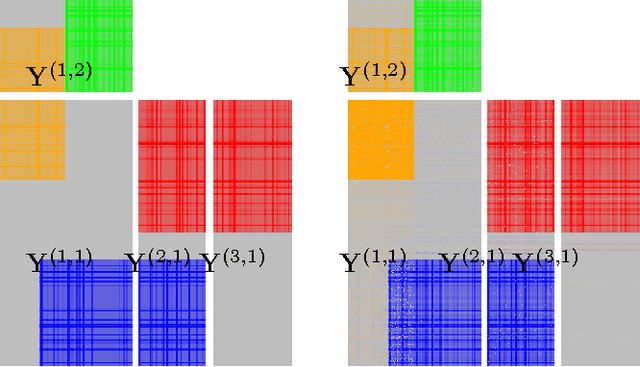


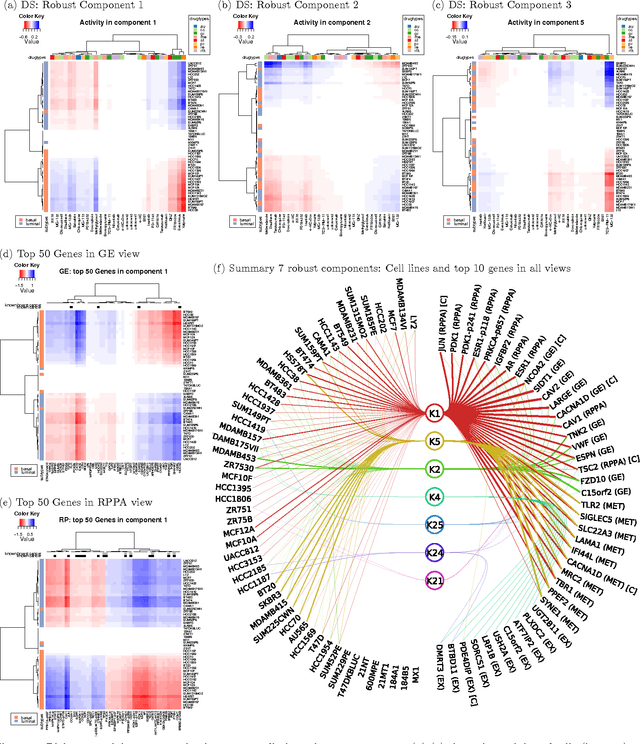
Abstract:Motivation: Modelling methods that find structure in data are necessary with the current large volumes of genomic data, and there have been various efforts to find subsets of genes exhibiting consistent patterns over subsets of treatments. These biclustering techniques have focused on one data source, often gene expression data. We present a Bayesian approach for joint biclustering of multiple data sources, extending a recent method Group Factor Analysis (GFA) to have a biclustering interpretation with additional sparsity assumptions. The resulting method enables data-driven detection of linear structure present in parts of the data sources. Results: Our simulation studies show that the proposed method reliably infers bi-clusters from heterogeneous data sources. We tested the method on data from the NCI-DREAM drug sensitivity prediction challenge, resulting in an excellent prediction accuracy. Moreover, the predictions are based on several biclusters which provide insight into the data sources, in this case on gene expression, DNA methylation, protein abundance, exome sequence, functional connectivity fingerprints and drug sensitivity.
* 7 pages, 5 figures, 1 table in Bioinformatics 2016
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge