Wiebke Arlt
Institute of Metabolism and Systems Research, University of Birmingham, the United Kingdom
Discriminative Subspace Emersion from learning feature relevances across different populations
Apr 02, 2025Abstract:In a given classification task, the accuracy of the learner is often hampered by finiteness of the training set, high-dimensionality of the feature space and severe overlap between classes. In the context of interpretable learners, with (piecewise) linear separation boundaries, these issues can be mitigated by careful construction of optimization procedures and/or estimation of relevant features for the task. However, when the task is shared across two disjoint populations the main interest is shifted towards estimating a set of features that discriminate the most between the two, when performing classification. We propose a new Discriminative Subspace Emersion (DSE) method to extend subspace learning toward a general relevance learning framework. DSE allows us to identify the most relevant features in distinguishing the classification task across two populations, even in cases of high overlap between classes. The proposed methodology is designed to work with multiple sets of labels and is derived in principle without being tied to a specific choice of base learner. Theoretical and empirical investigations over synthetic and real-world datasets indicate that DSE accurately identifies a common subspace for the classification across different populations. This is shown to be true for a surprisingly high degree of overlap between classes.
Interpretable Models Capable of Handling Systematic Missingness in Imbalanced Classes and Heterogeneous Datasets
Jun 04, 2022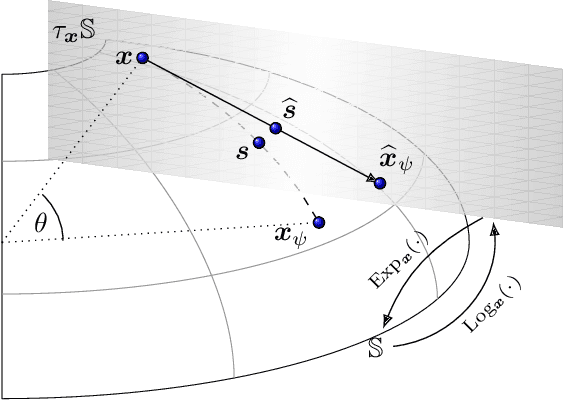
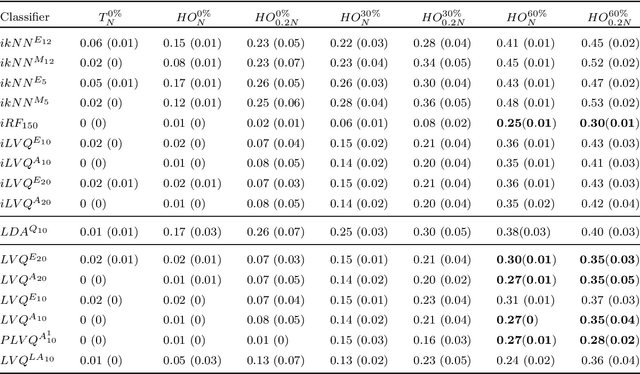

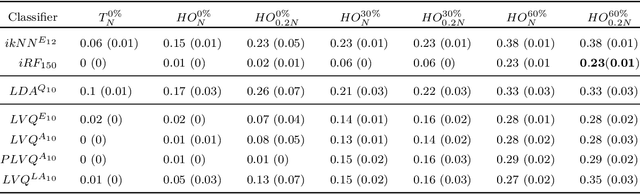
Abstract:Application of interpretable machine learning techniques on medical datasets facilitate early and fast diagnoses, along with getting deeper insight into the data. Furthermore, the transparency of these models increase trust among application domain experts. Medical datasets face common issues such as heterogeneous measurements, imbalanced classes with limited sample size, and missing data, which hinder the straightforward application of machine learning techniques. In this paper we present a family of prototype-based (PB) interpretable models which are capable of handling these issues. The models introduced in this contribution show comparable or superior performance to alternative techniques applicable in such situations. However, unlike ensemble based models, which have to compromise on easy interpretation, the PB models here do not. Moreover we propose a strategy of harnessing the power of ensembles while maintaining the intrinsic interpretability of the PB models, by averaging the model parameter manifolds. All the models were evaluated on a synthetic (publicly available dataset) in addition to detailed analyses of two real-world medical datasets (one publicly available). Results indicated that the models and strategies we introduced addressed the challenges of real-world medical data, while remaining computationally inexpensive and transparent, as well as similar or superior in performance compared to their alternatives.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge