Kelvin Lim
Imitation Learning with Limited Actions via Diffusion Planners and Deep Koopman Controllers
Oct 10, 2024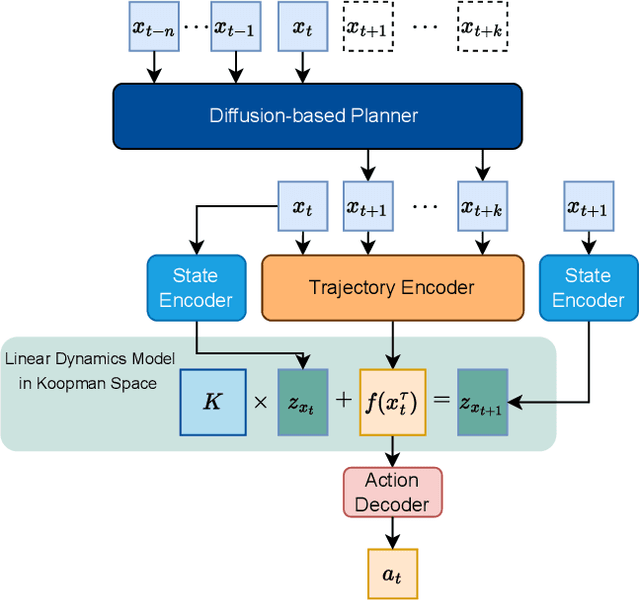
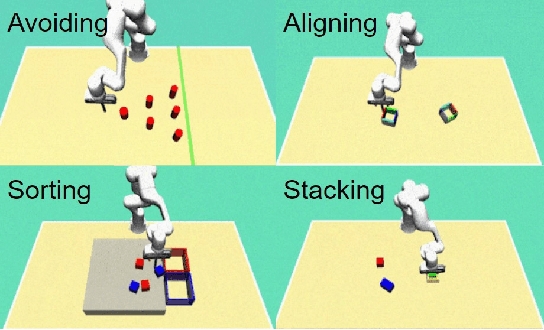
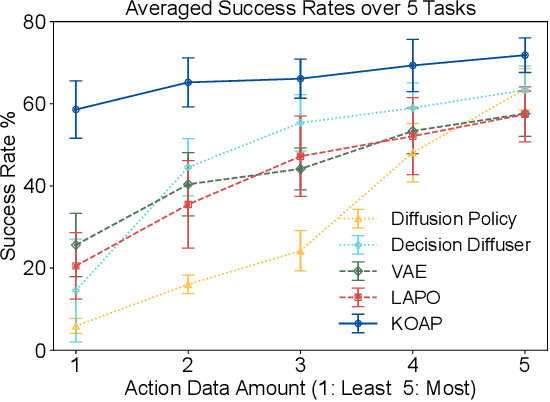
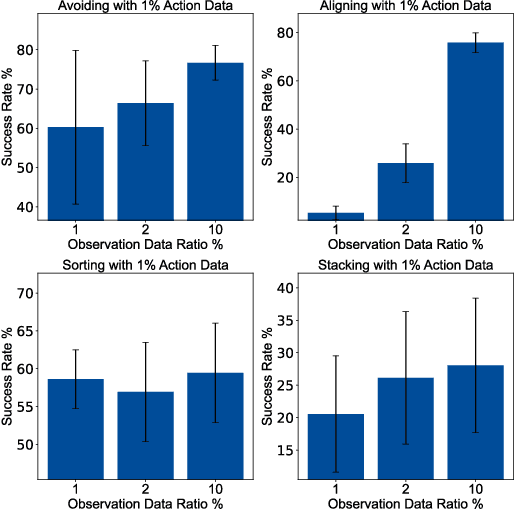
Abstract:Recent advances in diffusion-based robot policies have demonstrated significant potential in imitating multi-modal behaviors. However, these approaches typically require large quantities of demonstration data paired with corresponding robot action labels, creating a substantial data collection burden. In this work, we propose a plan-then-control framework aimed at improving the action-data efficiency of inverse dynamics controllers by leveraging observational demonstration data. Specifically, we adopt a Deep Koopman Operator framework to model the dynamical system and utilize observation-only trajectories to learn a latent action representation. This latent representation can then be effectively mapped to real high-dimensional continuous actions using a linear action decoder, requiring minimal action-labeled data. Through experiments on simulated robot manipulation tasks and a real robot experiment with multi-modal expert demonstrations, we demonstrate that our approach significantly enhances action-data efficiency and achieves high task success rates with limited action data.
Causal Discovery for fMRI data: Challenges, Solutions, and a Case Study
Dec 20, 2023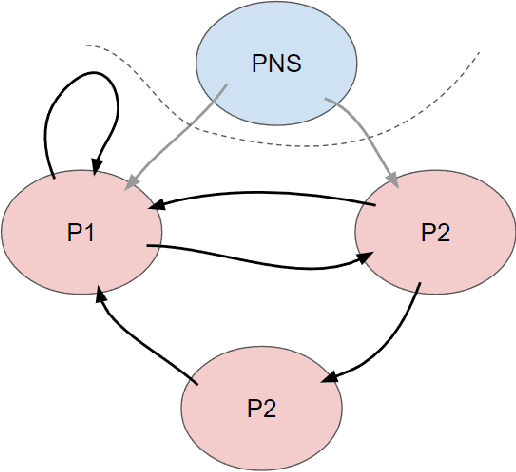
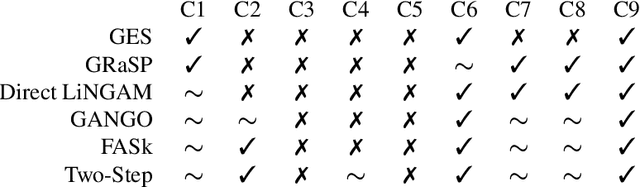
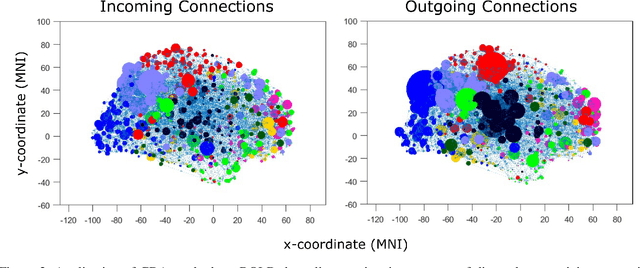
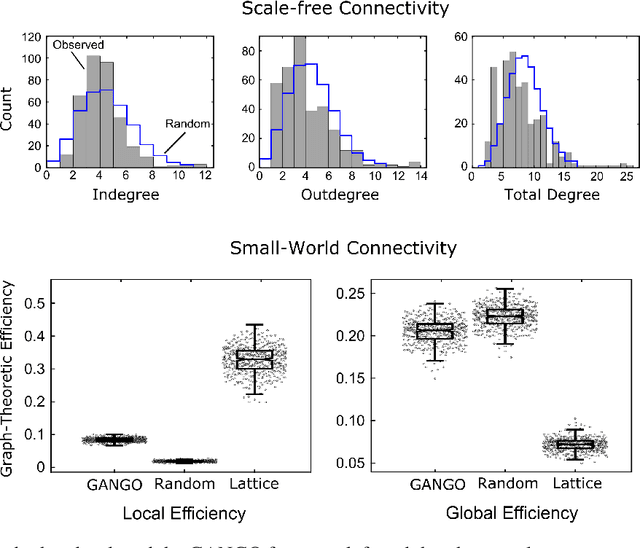
Abstract:Designing studies that apply causal discovery requires navigating many researcher degrees of freedom. This complexity is exacerbated when the study involves fMRI data. In this paper we (i) describe nine challenges that occur when applying causal discovery to fMRI data, (ii) discuss the space of decisions that need to be made, (iii) review how a recent case study made those decisions, (iv) and identify existing gaps that could potentially be solved by the development of new methods. Overall, causal discovery is a promising approach for analyzing fMRI data, and multiple successful applications have indicated that it is superior to traditional fMRI functional connectivity methods, but current causal discovery methods for fMRI leave room for improvement.
Penalized model-based clustering of fMRI data
Oct 13, 2020



Abstract:Functional magnetic resonance imaging (fMRI) data have become increasingly available and are useful for describing functional connectivity (FC), the relatedness of neuronal activity in regions of the brain. This FC of the brain provides insight into certain neurodegenerative diseases and psychiatric disorders, and thus is of clinical importance. To help inform physicians regarding patient diagnoses, unsupervised clustering of subjects based on FC is desired, allowing the data to inform us of groupings of patients based on shared features of connectivity. Since heterogeneity in FC is present even between patients within the same group, it is important to allow subject-level differences in connectivity, while still pooling information across patients within each group to describe group-level FC. To this end, we propose a random covariance clustering model (RCCM) to concurrently cluster subjects based on their FC networks, estimate the unique FC networks of each subject, and to infer shared network features. Although current methods exist for estimating FC or clustering subjects using fMRI data, our novel contribution is to cluster or group subjects based on similar FC of the brain while simultaneously providing group- and subject-level FC network estimates. The competitive performance of RCCM relative to other methods is demonstrated through simulations in various settings, achieving both improved clustering of subjects and estimation of FC networks. Utility of the proposed method is demonstrated with application to a resting-state fMRI data set collected on 43 healthy controls and 61 participants diagnosed with schizophrenia.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge