Juraj Vladika
Facts Fade Fast: Evaluating Memorization of Outdated Medical Knowledge in Large Language Models
Sep 04, 2025Abstract:The growing capabilities of Large Language Models (LLMs) show significant potential to enhance healthcare by assisting medical researchers and physicians. However, their reliance on static training data is a major risk when medical recommendations evolve with new research and developments. When LLMs memorize outdated medical knowledge, they can provide harmful advice or fail at clinical reasoning tasks. To investigate this problem, we introduce two novel question-answering (QA) datasets derived from systematic reviews: MedRevQA (16,501 QA pairs covering general biomedical knowledge) and MedChangeQA (a subset of 512 QA pairs where medical consensus has changed over time). Our evaluation of eight prominent LLMs on the datasets reveals consistent reliance on outdated knowledge across all models. We additionally analyze the influence of obsolete pre-training data and training strategies to explain this phenomenon and propose future directions for mitigation, laying the groundwork for developing more current and reliable medical AI systems.
Correcting Hallucinations in News Summaries: Exploration of Self-Correcting LLM Methods with External Knowledge
Jun 24, 2025Abstract:While large language models (LLMs) have shown remarkable capabilities to generate coherent text, they suffer from the issue of hallucinations -- factually inaccurate statements. Among numerous approaches to tackle hallucinations, especially promising are the self-correcting methods. They leverage the multi-turn nature of LLMs to iteratively generate verification questions inquiring additional evidence, answer them with internal or external knowledge, and use that to refine the original response with the new corrections. These methods have been explored for encyclopedic generation, but less so for domains like news summarization. In this work, we investigate two state-of-the-art self-correcting systems by applying them to correct hallucinated summaries using evidence from three search engines. We analyze the results and provide insights into systems' performance, revealing interesting practical findings on the benefits of search engine snippets and few-shot prompts, as well as high alignment of G-Eval and human evaluation.
Improving Reliability and Explainability of Medical Question Answering through Atomic Fact Checking in Retrieval-Augmented LLMs
May 30, 2025Abstract:Large language models (LLMs) exhibit extensive medical knowledge but are prone to hallucinations and inaccurate citations, which pose a challenge to their clinical adoption and regulatory compliance. Current methods, such as Retrieval Augmented Generation, partially address these issues by grounding answers in source documents, but hallucinations and low fact-level explainability persist. In this work, we introduce a novel atomic fact-checking framework designed to enhance the reliability and explainability of LLMs used in medical long-form question answering. This method decomposes LLM-generated responses into discrete, verifiable units called atomic facts, each of which is independently verified against an authoritative knowledge base of medical guidelines. This approach enables targeted correction of errors and direct tracing to source literature, thereby improving the factual accuracy and explainability of medical Q&A. Extensive evaluation using multi-reader assessments by medical experts and an automated open Q&A benchmark demonstrated significant improvements in factual accuracy and explainability. Our framework achieved up to a 40% overall answer improvement and a 50% hallucination detection rate. The ability to trace each atomic fact back to the most relevant chunks from the database provides a granular, transparent explanation of the generated responses, addressing a major gap in current medical AI applications. This work represents a crucial step towards more trustworthy and reliable clinical applications of LLMs, addressing key prerequisites for clinical application and fostering greater confidence in AI-assisted healthcare.
On the Influence of Context Size and Model Choice in Retrieval-Augmented Generation Systems
Feb 20, 2025



Abstract:Retrieval-augmented generation (RAG) has emerged as an approach to augment large language models (LLMs) by reducing their reliance on static knowledge and improving answer factuality. RAG retrieves relevant context snippets and generates an answer based on them. Despite its increasing industrial adoption, systematic exploration of RAG components is lacking, particularly regarding the ideal size of provided context, and the choice of base LLM and retrieval method. To help guide development of robust RAG systems, we evaluate various context sizes, BM25 and semantic search as retrievers, and eight base LLMs. Moving away from the usual RAG evaluation with short answers, we explore the more challenging long-form question answering in two domains, where a good answer has to utilize the entire context. Our findings indicate that final QA performance improves steadily with up to 15 snippets but stagnates or declines beyond that. Finally, we show that different general-purpose LLMs excel in the biomedical domain than the encyclopedic one, and that open-domain evidence retrieval in large corpora is challenging.
Step-by-Step Fact Verification System for Medical Claims with Explainable Reasoning
Feb 20, 2025Abstract:Fact verification (FV) aims to assess the veracity of a claim based on relevant evidence. The traditional approach for automated FV includes a three-part pipeline relying on short evidence snippets and encoder-only inference models. More recent approaches leverage the multi-turn nature of LLMs to address FV as a step-by-step problem where questions inquiring additional context are generated and answered until there is enough information to make a decision. This iterative method makes the verification process rational and explainable. While these methods have been tested for encyclopedic claims, exploration on domain-specific and realistic claims is missing. In this work, we apply an iterative FV system on three medical fact-checking datasets and evaluate it with multiple settings, including different LLMs, external web search, and structured reasoning using logic predicates. We demonstrate improvements in the final performance over traditional approaches and the high potential of step-by-step FV systems for domain-specific claims.
Lexical Substitution is not Synonym Substitution: On the Importance of Producing Contextually Relevant Word Substitutes
Feb 06, 2025Abstract:Lexical Substitution is the task of replacing a single word in a sentence with a similar one. This should ideally be one that is not necessarily only synonymous, but also fits well into the surrounding context of the target word, while preserving the sentence's grammatical structure. Recent advances in Lexical Substitution have leveraged the masked token prediction task of Pre-trained Language Models to generate replacements for a given word in a sentence. With this technique, we introduce ConCat, a simple augmented approach which utilizes the original sentence to bolster contextual information sent to the model. Compared to existing approaches, it proves to be very effective in guiding the model to make contextually relevant predictions for the target word. Our study includes a quantitative evaluation, measured via sentence similarity and task performance. In addition, we conduct a qualitative human analysis to validate that users prefer the substitutions proposed by our method, as opposed to previous methods. Finally, we test our approach on the prevailing benchmark for Lexical Substitution, CoInCo, revealing potential pitfalls of the benchmark. These insights serve as the foundation for a critical discussion on the way in which Lexical Substitution is evaluated.
Adapter-based Approaches to Knowledge-enhanced Language Models -- A Survey
Nov 25, 2024



Abstract:Knowledge-enhanced language models (KELMs) have emerged as promising tools to bridge the gap between large-scale language models and domain-specific knowledge. KELMs can achieve higher factual accuracy and mitigate hallucinations by leveraging knowledge graphs (KGs). They are frequently combined with adapter modules to reduce the computational load and risk of catastrophic forgetting. In this paper, we conduct a systematic literature review (SLR) on adapter-based approaches to KELMs. We provide a structured overview of existing methodologies in the field through quantitative and qualitative analysis and explore the strengths and potential shortcomings of individual approaches. We show that general knowledge and domain-specific approaches have been frequently explored along with various adapter architectures and downstream tasks. We particularly focused on the popular biomedical domain, where we provided an insightful performance comparison of existing KELMs. We outline the main trends and propose promising future directions.
* 12 pages, 4 figures. Published at KEOD24 via SciTePress
Towards Optimizing a Retrieval Augmented Generation using Large Language Model on Academic Data
Nov 13, 2024



Abstract:Given the growing trend of many organizations integrating Retrieval Augmented Generation (RAG) into their operations, we assess RAG on domain-specific data and test state-of-the-art models across various optimization techniques. We incorporate four optimizations; Multi-Query, Child-Parent-Retriever, Ensemble Retriever, and In-Context-Learning, to enhance the functionality and performance in the academic domain. We focus on data retrieval, specifically targeting various study programs at a large technical university. We additionally introduce a novel evaluation approach, the RAG Confusion Matrix designed to assess the effectiveness of various configurations within the RAG framework. By exploring the integration of both open-source (e.g., Llama2, Mistral) and closed-source (GPT-3.5 and GPT-4) Large Language Models, we offer valuable insights into the application and optimization of RAG frameworks in domain-specific contexts. Our experiments show a significant performance increase when including multi-query in the retrieval phase.
Enhancing Answer Attribution for Faithful Text Generation with Large Language Models
Oct 22, 2024



Abstract:The increasing popularity of Large Language Models (LLMs) in recent years has changed the way users interact with and pose questions to AI-based conversational systems. An essential aspect for increasing the trustworthiness of generated LLM answers is the ability to trace the individual claims from responses back to relevant sources that support them, the process known as answer attribution. While recent work has started exploring the task of answer attribution in LLMs, some challenges still remain. In this work, we first perform a case study analyzing the effectiveness of existing answer attribution methods, with a focus on subtasks of answer segmentation and evidence retrieval. Based on the observed shortcomings, we propose new methods for producing more independent and contextualized claims for better retrieval and attribution. The new methods are evaluated and shown to improve the performance of answer attribution components. We end with a discussion and outline of future directions for the task.
DP-MLM: Differentially Private Text Rewriting Using Masked Language Models
Jun 30, 2024
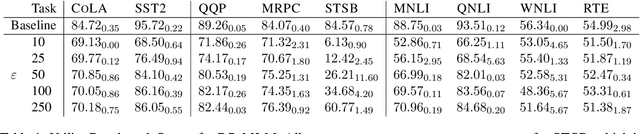
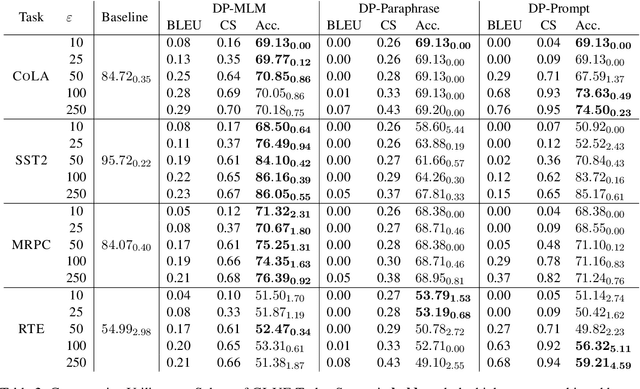
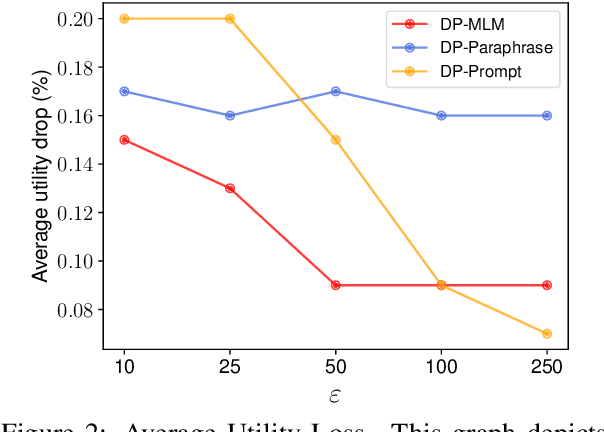
Abstract:The task of text privatization using Differential Privacy has recently taken the form of $\textit{text rewriting}$, in which an input text is obfuscated via the use of generative (large) language models. While these methods have shown promising results in the ability to preserve privacy, these methods rely on autoregressive models which lack a mechanism to contextualize the private rewriting process. In response to this, we propose $\textbf{DP-MLM}$, a new method for differentially private text rewriting based on leveraging masked language models (MLMs) to rewrite text in a semantically similar $\textit{and}$ obfuscated manner. We accomplish this with a simple contextualization technique, whereby we rewrite a text one token at a time. We find that utilizing encoder-only MLMs provides better utility preservation at lower $\varepsilon$ levels, as compared to previous methods relying on larger models with a decoder. In addition, MLMs allow for greater customization of the rewriting mechanism, as opposed to generative approaches. We make the code for $\textbf{DP-MLM}$ public and reusable, found at https://github.com/sjmeis/DPMLM .
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge