Jiazhen Chen
Towards Multiple Missing Values-resistant Unsupervised Graph Anomaly Detection
Nov 13, 2025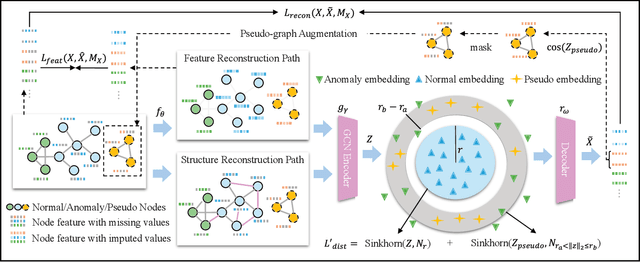
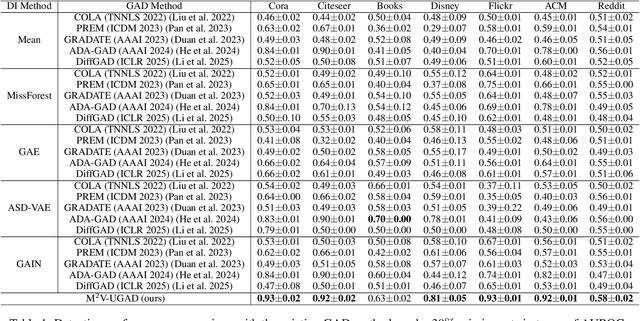
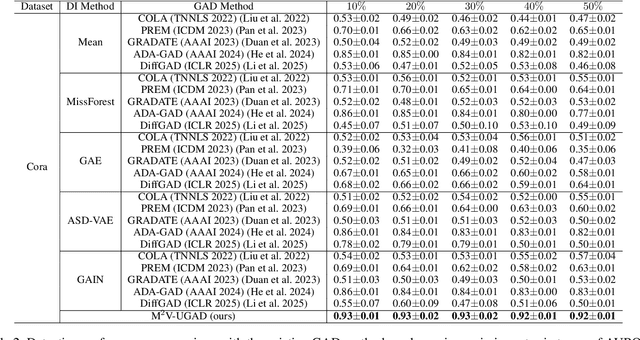
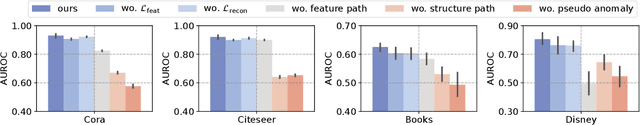
Abstract:Unsupervised graph anomaly detection (GAD) has received increasing attention in recent years, which aims to identify data anomalous patterns utilizing only unlabeled node information from graph-structured data. However, prevailing unsupervised GAD methods typically presuppose complete node attributes and structure information, a condition hardly satisfied in real-world scenarios owing to privacy, collection errors or dynamic node arrivals. Existing standard imputation schemes risk "repairing" rare anomalous nodes so that they appear normal, thereby introducing imputation bias into the detection process. In addition, when both node attributes and edges are missing simultaneously, estimation errors in one view can contaminate the other, causing cross-view interference that further undermines the detection performance. To overcome these challenges, we propose M$^2$V-UGAD, a multiple missing values-resistant unsupervised GAD framework on incomplete graphs. Specifically, a dual-pathway encoder is first proposed to independently reconstruct missing node attributes and graph structure, thereby preventing errors in one view from propagating to the other. The two pathways are then fused and regularized in a joint latent space so that normals occupy a compact inner manifold while anomalies reside on an outer shell. Lastly, to mitigate imputation bias, we sample latent codes just outside the normal region and decode them into realistic node features and subgraphs, providing hard negative examples that sharpen the decision boundary. Experiments on seven public benchmarks demonstrate that M$^2$V-UGAD consistently outperforms existing unsupervised GAD methods across varying missing rates.
Towards Effective Open-set Graph Class-incremental Learning
Jul 23, 2025Abstract:Graph class-incremental learning (GCIL) allows graph neural networks (GNNs) to adapt to evolving graph analytical tasks by incrementally learning new class knowledge while retaining knowledge of old classes. Existing GCIL methods primarily focus on a closed-set assumption, where all test samples are presumed to belong to previously known classes. Such an assumption restricts their applicability in real-world scenarios, where unknown classes naturally emerge during inference, and are absent during training. In this paper, we explore a more challenging open-set graph class-incremental learning scenario with two intertwined challenges: catastrophic forgetting of old classes, which impairs the detection of unknown classes, and inadequate open-set recognition, which destabilizes the retention of learned knowledge. To address the above problems, a novel OGCIL framework is proposed, which utilizes pseudo-sample embedding generation to effectively mitigate catastrophic forgetting and enable robust detection of unknown classes. To be specific, a prototypical conditional variational autoencoder is designed to synthesize node embeddings for old classes, enabling knowledge replay without storing raw graph data. To handle unknown classes, we employ a mixing-based strategy to generate out-of-distribution (OOD) samples from pseudo in-distribution and current node embeddings. A novel prototypical hypersphere classification loss is further proposed, which anchors in-distribution embeddings to their respective class prototypes, while repelling OOD embeddings away. Instead of assigning all unknown samples into one cluster, our proposed objective function explicitly models them as outliers through prototype-aware rejection regions, ensuring a robust open-set recognition. Extensive experiments on five benchmarks demonstrate the effectiveness of OGCIL over existing GCIL and open-set GNN methods.
Semi-supervised Anomaly Detection with Extremely Limited Labels in Dynamic Graphs
Jan 25, 2025Abstract:Semi-supervised graph anomaly detection (GAD) has recently received increasing attention, which aims to distinguish anomalous patterns from graphs under the guidance of a moderate amount of labeled data and a large volume of unlabeled data. Although these proposed semi-supervised GAD methods have achieved great success, their superior performance will be seriously degraded when the provided labels are extremely limited due to some unpredictable factors. Besides, the existing methods primarily focus on anomaly detection in static graphs, and little effort was paid to consider the continuous evolution characteristic of graphs over time (dynamic graphs). To address these challenges, we propose a novel GAD framework (EL$^{2}$-DGAD) to tackle anomaly detection problem in dynamic graphs with extremely limited labels. Specifically, a transformer-based graph encoder model is designed to more effectively preserve evolving graph structures beyond the local neighborhood. Then, we incorporate an ego-context hypersphere classification loss to classify temporal interactions according to their structure and temporal neighborhoods while ensuring the normal samples are mapped compactly against anomalous data. Finally, the above loss is further augmented with an ego-context contrasting module which utilizes unlabeled data to enhance model generalization. Extensive experiments on four datasets and three label rates demonstrate the effectiveness of the proposed method in comparison to the existing GAD methods.
Disentangling the Complex Multiplexed DIA Spectra in De Novo Peptide Sequencing
Nov 24, 2024Abstract:Data-Independent Acquisition (DIA) was introduced to improve sensitivity to cover all peptides in a range rather than only sampling high-intensity peaks as in Data-Dependent Acquisition (DDA) mass spectrometry. However, it is not very clear how useful DIA data is for de novo peptide sequencing as the DIA data are marred with coeluted peptides, high noises, and varying data quality. We present a new deep learning method DIANovo, and address each of these difficulties, and improves the previous established system DeepNovo-DIA by from 25% to 81%, averaging 48%, for amino acid recall, and by from 27% to 89%, averaging 57%, for peptide recall, by equipping the model with a deeper understanding of coeluted DIA spectra. This paper also provides criteria about when DIA data could be used for de novo peptide sequencing and when not to by providing a comparison between DDA and DIA, in both de novo and database search mode. We find that while DIA excels with narrow isolation windows on older-generation instruments, it loses its advantage with wider windows. However, with Orbitrap Astral, DIA consistently outperforms DDA due to narrow window mode enabled. We also provide a theoretical explanation of this phenomenon, emphasizing the critical role of the signal-to-noise profile in the successful application of de novo sequencing.
Towards Cross-domain Few-shot Graph Anomaly Detection
Oct 11, 2024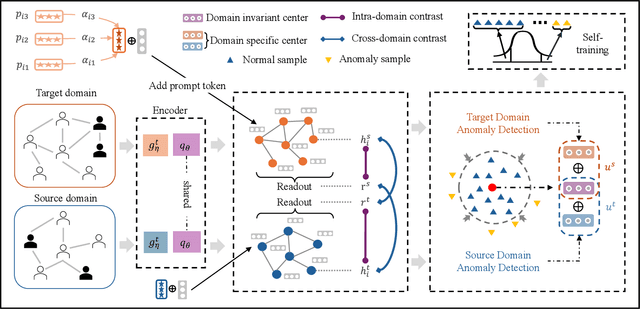
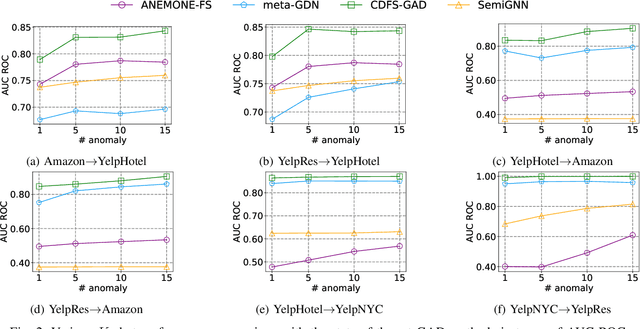
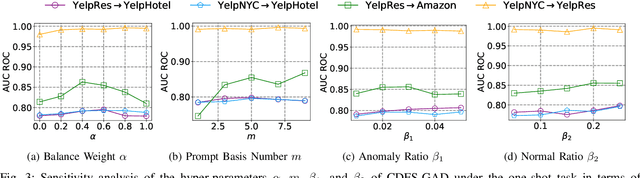
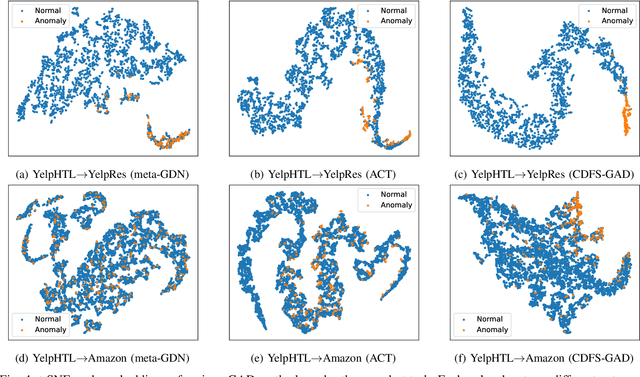
Abstract:Few-shot graph anomaly detection (GAD) has recently garnered increasing attention, which aims to discern anomalous patterns among abundant unlabeled test nodes under the guidance of a limited number of labeled training nodes. Existing few-shot GAD approaches typically adopt meta-training methods trained on richly labeled auxiliary networks to facilitate rapid adaptation to target networks that possess sparse labels. However, these proposed methods often assume that the auxiliary and target networks exist in the same data distributions-an assumption rarely holds in practical settings. This paper explores a more prevalent and complex scenario of cross-domain few-shot GAD, where the goal is to identify anomalies within sparsely labeled target graphs using auxiliary graphs from a related, yet distinct domain. The challenge here is nontrivial owing to inherent data distribution discrepancies between the source and target domains, compounded by the uncertainties of sparse labeling in the target domain. In this paper, we propose a simple and effective framework, termed CDFS-GAD, specifically designed to tackle the aforementioned challenges. CDFS-GAD first introduces a domain-adaptive graph contrastive learning module, which is aimed at enhancing cross-domain feature alignment. Then, a prompt tuning module is further designed to extract domain-specific features tailored to each domain. Moreover, a domain-adaptive hypersphere classification loss is proposed to enhance the discrimination between normal and anomalous instances under minimal supervision, utilizing domain-sensitive norms. Lastly, a self-training strategy is introduced to further refine the predicted scores, enhancing its reliability in few-shot settings. Extensive experiments on twelve real-world cross-domain data pairs demonstrate the effectiveness of the proposed CDFS-GAD framework in comparison to various existing GAD methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge