Jiaxin Zheng
The NLP Sandbox: an efficient model-to-data system to enable federated and unbiased evaluation of clinical NLP models
Jun 28, 2022



Abstract:Objective The evaluation of natural language processing (NLP) models for clinical text de-identification relies on the availability of clinical notes, which is often restricted due to privacy concerns. The NLP Sandbox is an approach for alleviating the lack of data and evaluation frameworks for NLP models by adopting a federated, model-to-data approach. This enables unbiased federated model evaluation without the need for sharing sensitive data from multiple institutions. Materials and Methods We leveraged the Synapse collaborative framework, containerization software, and OpenAPI generator to build the NLP Sandbox (nlpsandbox.io). We evaluated two state-of-the-art NLP de-identification focused annotation models, Philter and NeuroNER, using data from three institutions. We further validated model performance using data from an external validation site. Results We demonstrated the usefulness of the NLP Sandbox through de-identification clinical model evaluation. The external developer was able to incorporate their model into the NLP Sandbox template and provide user experience feedback. Discussion We demonstrated the feasibility of using the NLP Sandbox to conduct a multi-site evaluation of clinical text de-identification models without the sharing of data. Standardized model and data schemas enable smooth model transfer and implementation. To generalize the NLP Sandbox, work is required on the part of data owners and model developers to develop suitable and standardized schemas and to adapt their data or model to fit the schemas. Conclusions The NLP Sandbox lowers the barrier to utilizing clinical data for NLP model evaluation and facilitates federated, multi-site, unbiased evaluation of NLP models.
The RSNA-ASNR-MICCAI BraTS 2021 Benchmark on Brain Tumor Segmentation and Radiogenomic Classification
Jul 05, 2021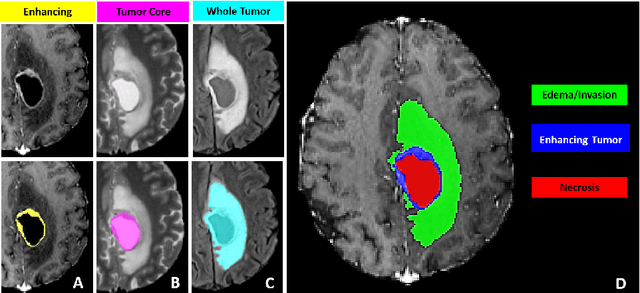
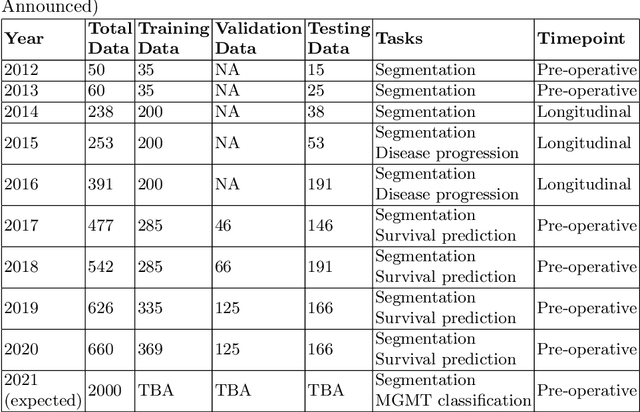
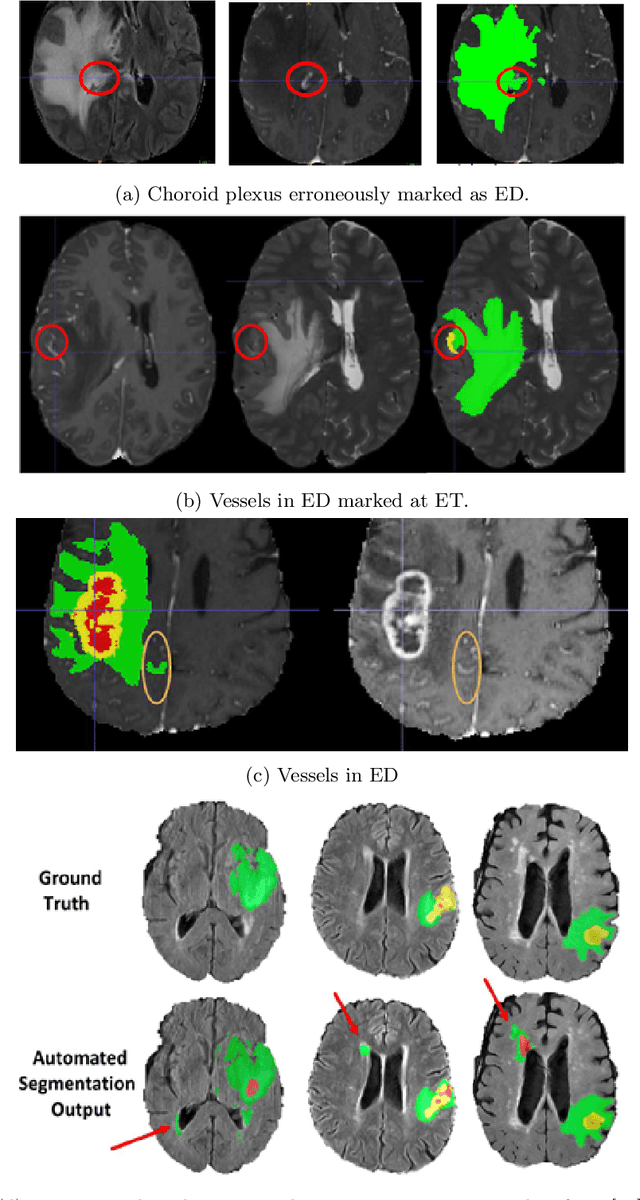
Abstract:The BraTS 2021 challenge celebrates its 10th anniversary and is jointly organized by the Radiological Society of North America (RSNA), the American Society of Neuroradiology (ASNR), and the Medical Image Computing and Computer Assisted Interventions (MICCAI) society. Since its inception, BraTS has been focusing on being a common benchmarking venue for brain glioma segmentation algorithms, with well-curated multi-institutional multi-parametric magnetic resonance imaging (mpMRI) data. Gliomas are the most common primary malignancies of the central nervous system, with varying degrees of aggressiveness and prognosis. The RSNA-ASNR-MICCAI BraTS 2021 challenge targets the evaluation of computational algorithms assessing the same tumor compartmentalization, as well as the underlying tumor's molecular characterization, in pre-operative baseline mpMRI data from 2,000 patients. Specifically, the two tasks that BraTS 2021 focuses on are: a) the segmentation of the histologically distinct brain tumor sub-regions, and b) the classification of the tumor's O[6]-methylguanine-DNA methyltransferase (MGMT) promoter methylation status. The performance evaluation of all participating algorithms in BraTS 2021 will be conducted through the Sage Bionetworks Synapse platform (Task 1) and Kaggle (Task 2), concluding in distributing to the top ranked participants monetary awards of $60,000 collectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge